Introduction
Materials and Methods
Taxon Sampling, Genome Sequencing, Chloroplast Genome Assembly and Annotation
Drawing circular map of chloroplast genome
Comparative analysis of chloroplast genomes
Simple sequence repeat (SSR) analysis
Nucleotide diversity analysis
Phylogenetic analysis
Results and Discussion
Chloroplast genome features
Simple sequence repeats analyses of Dysphania chloroplast genomes
Nucleotide diversity analysis within three Dysphania species
Comparative analysis of IR junction structure
Phylogenetic analyses with complete chloroplast genomes
Conclusion
Introduction
Genus Dysphania R. Br. (Chenopodiaceae/Amaranthaceae sensu in APG system) contained only ten species originated from Australia (Wilson et al., 1983), but now this genus is composed of 48 taxa by including species from other genera (Mosyakin and Clemants, 2002; Mosyakin and Clemants, 2008; Perth, 2011; Verloove and Lambinon, 2006) as well as newly reported species in Chenopodiaceae (Dillon and Markey, 2016; Sukhorukov, 2012; Sukhorukov et al., 2015). This genus is characterized by presence of multicellular glandular hair in Flora of China (Zhu et al., 2003). The phylogenetic position of genus Dysphania have been confirmed and maintained as a robust monophyletic clade supported by many phylogenetic studies (Fuentes-Bazan et al., 2012a; Fuentes-Bazan et al., 2012b; Kadereit et al., 2003; Kadereit et al., 2010). Thanks to rapid development of next generation sequencing (NGS) technologies (Mardis, 2008; Metzker, 2010), several complete chloroplast genomes of Dysphania species have been sequenced to understand their genetic information as a candidate of cancer treatment (Chen and Yang, 2018), an invasive species (Kim et al., 2019), and investigation of intraspecies variation (Park and Kim, 2019).
Dysphania ambrosioides (L.) Mosyakin & Clemants (= Chenopodium ambrosioides L.) is South American ethnobotanical species utilized as a medicinal and culinary plant named as i) ‘epazote’ by Aztecs and Mayans in Mesoamerica, ii) ‘paico’ in Andean region, and many others names of South America (Albuquerque et al., 2018). It is native to Central and Southern America and is now distributed throughout most of the world including North America (Stohlgren et al., 2013), Europe (Balogh et al., 2003; Goia et al., 2014; Maslo, 2016; Pace and Tammaro, 2001; Weber and Gut, 2004), Asia (Aravindhan and Rajendran, 2014; Dogra et al., 2009; Liu et al., 2006; Mito and Uesugi, 2004; Rastogi et al., 2015; Sekar, 2012; Xu et al., 2012), and Africa (Brown et al., 1985; Foxcroft et al., 2008; Hegazy et al., 2008; Verloove, 2013). D. ambrosioides was introduced and naturalized in South Korea (Han et al., 2018; Lee et al., 2011). Until now, there is no report of agricultural and ecological damages where D. amrosioides has been found, but it is possible to affect harmful effects to local ecosystems; e.g., volatile oil of D. ambrosioides inhibits germination and seeding growth of rape (Brassica campestris L.), lettuce (Lactuca sativa L.), and wheat (Triticum aestivum L.) due to its allelopathic effect (Wang et al., 2009).
Despite its negative effects, useful effects of D. ambrosioides have been reported and confirmed; for example, anti-inflammatory (Ibironke and Ajiboye, 2007; Reyes-Becerril et al., 2019), antioxidant (Bezerra et al., 2019; Kumar et al., 2007), anti-tumoral action and immunostimulatory (Rossi-Bergamann et al., 1997), antifungal (Chekem et al., 2010; Jardim et al., 2008; Kumar et al., 2007; Prasad et al., 2010), antimicrobial (Bezerra et al., 2019; Brahim et al., 2015; Kiuchi et al., 2002; Monzote et al., 2006; Monzote et al., 2014), insecticidal (Pandey et al., 2014; Pavela et al., 2018; Wei et al., 2015), and even corrosion inhibition of steel (Bammou et al., 2014). These positive effects led us to decipher its chloroplast genome to understand genetic background and phylogenetic position under genus Dysphania. It was also analyzed to understand phylogenomic position and to compare the characteristics of inter- and intra-species among Dysphania species.
Materials and Methods
Taxon Sampling, Genome Sequencing, Chloroplast Genome Assembly and Annotation
Fresh Dysphania ambrosioides leaves from a single individual were collected from Korea (voucher specimen: IB-01027, Y. Kim, in InfoBoss Cyber Herbarium (IN)). Total DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA) and sequenced using the Illumina HiSeqX (Illumina, Inc., San Diego, CA, USA) at Macrogen Corporation (Seoul, Korea). Raw sequences were filtered by Trimmomatic 0.33 (Bolger et al., 2014). The resulting paired-end reads were assembled de novo using Velvet 1.2.10 (Zerbino and Birney, 2008) with multiple k-mers ranging from 51 to 81 to select the best assembly result. Gap filling process was conducted with SOAPGapCloser 1.12 (Zhao et al., 2011). Assembled sequences were confirmed using BWA 0.7.17 (Li, 2013) and SAMtools 1.9 (Li et al., 2009) to correct misassembled bases. The tRNAs were confirmed using tRNAscan-SE (Lowe and Eddy, 1997). Annotation was conducted using Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) with Dysphania pumilio chloroplast genome (NC_041159; Kim et al., 2019), and the annotated chloroplast genome sequences were submitted to GenBank (accession number is NC_041201).
Drawing circular map of chloroplast genome
The annotated GenBank format sequence file was used to draw the circular map using OGDRAW 1.2 (Lohse et al., 2007) with default options.
Comparative analysis of chloroplast genomes
The complete chloroplast of D. ambrosioides was compared to those of the other available Dysphania species including two D. pumilio and one D. botrys by aligning these sequences with MAFFT 7.388 (Katoh and Standley, 2013). Based on these alignment, nucleotide diversity was calcualted by inhouse pipeline implemented in the PCD (http://www.cp-genome.net; Park et al., in preparation). Calculation was done with parameters: 500-bp windows and 20-bp steps.
Simple sequence repeat (SSR) analysis
Simple sequence repeats (SSRs) were identified on the chloroplast genome sequence using the pipeline of the SSR database (SSRDB; http://ssr.pe.kr/; Park et al., in preparation). Based on conventional definition of SSR on chloroplast genome: MonoSSR (1 bp) to HexaSSR (6 bp) and total length of SSRs on chloroplast genome is above 10 bp. However, practically, many researches used on additional condition: number of minimum repeats of MonoSSR is 8 not 10 (Chen et al., 2015), and minimum repeats of PentaSSR and HexaSSR are 3 (Jeon and Kim, 2019; Li et al., 2019; Shukla et al., 2018) or 4 (Cheng et al., 2016; Kim et al., 2019) instead of 2. It may be caused by the reason that once there is sequence variation in SSR of which repeat is 2 and then it is no longer SSR because number of repeats of this mutated SSR is only 1. In addition, one research used one condition that number of repeats of MonoSSR can be 9 when there is corresponding SSR of which number of repeats is 10 in other species (Jeon and Kim, 2019).
So, we made the rule to identify SSRs on chloroplast genomes like this: MonoSSR is defined that unit sequence length is 1 bp and number of repeats is at least 10, DiSSR is defined that unit sequence length is 2 bp and number of repeats is at least 5, TriSSR is defined that unit sequence length is 3 bp and number of repeats is at least 4, and TetraSSR is defined that unit sequence length is 4 bp and number of repeats is at least 3 to keep minimum length of chloroplast SSR (10 bp). For PentaSSR and HexaSSR, one additional group was defined named as potential SSRs: PentaSSRs and HexaSSRs of which number of repeats is 2. In addition, we extended range of SSRs of which motif length is 7 bp to 10 bp with minimum two repeats named as HeptaSSR, OctaSSR, NonaSSR, and DecaSSR.
After identifying SSRs on chloroplast genomes, their coordinates were compared with gene position to classify where they are originated from under the SSRDB (http://ssr.pe.kr/).
Nucleotide diversity analysis
Nucleotide diversity was calculated based on the method proposed by Nei and Li (1979) based on multiple sequence alignment of three Dysphania chloroplast genome sequences using perl script. Window size is 500 bp and step size is 200 bp for sliding-window method. Genomic coordination of each window was compared with gene annotation of chloroplast genome for further analyses.
Phylogenetic analysis
For the phylogenetic analysis, complete chloroplast genomes of 26 representative species from the Chenopodiaceae/Amaranthaceae sensu in APG system and seven species as outgroup species were aligned using MAFFT 7.388 (Katoh and Standley, 2013). All chloroplast genome sequences were retrieved from the PCD (http://www.cp-genome.net; Park et al., in preparation). The maximum likelihood (ML) phylogeny tree was constructed by IQ-TREE 1.6.6 (Nguyen et al., 2014) under the GTR+F+R3 model (1,000 replicates) and phylogenetic tree was modified using FigTree 1.4.4 (http://tree. bio.ed.ac.uk/software/figtree/).
Results and Discussion
Chloroplast genome features
Chloroplast genome of Dysphaniaambrosioides (GenBank accession is NC_041201) is 151,689 bp long and has four subregions: 83,421 bp of large single copy (LSC) and 18,062 bp of small single copy (SSC) regions are separated by 25,103 bp of inverted repeat (IR). GC ratio of this chloroplast genome is 36.9% and those of LSC, SSC, and IR are 34.9%, 30.3%, and 42.7%, respectively. It contains 128 genes (84 protein-coding genes, eight rRNAs, and 36 tRNAs); 15 genes including seven tRNA genes (trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA, and trnI-CAU), four rRNA genes (rRNA5, rRNA4.5, rRNA23, and rRNA16), and four protein-coding genes (rps7, ndhB, ycf2, and rpl2) are duplicated in IR regions. Twelve genes (ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, atpF, trnI-GAU, trnA-UGC, trnK-UUU, and trnL-UAA) contain one intron, while clpP, rps12, and ycf3 have two introns. In addition, the complete ycf1 gene is located in the IR region at the SSC/IR junction (Fig. 1). The number of genes and gene order are identical in the chloroplast genomes of three Dysphania species except GC ratio (Table 1). The GC ratio among those species is slightly diffident in LSC (34.7% to 34.9%) and SSC (30.1% to 30.4%), while that of IR is same to each other (Table 1).
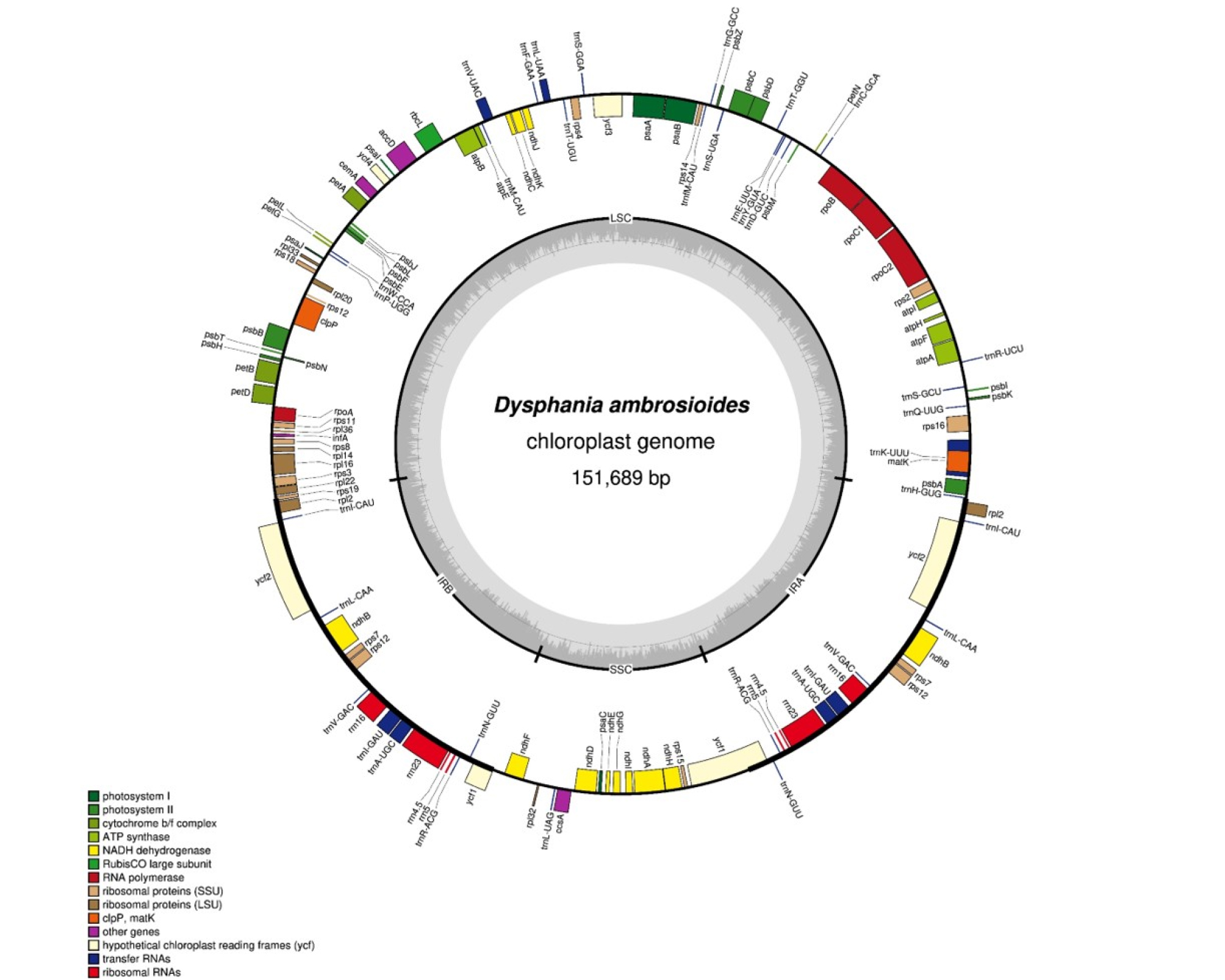
Fig. 1.
Gene map of Dysphania ambrosioides chloroplast genome. The genes located outside of the circle are transcribed clockwise, while those located inside are transcribed counterclockwise. The dark gray plot in the inner circle corresponds to GC content. Large single copy, small single copy, and inverted repeat are indicated with LSC, SSC, and IR (IRa and IRb), respectively.
Table 1. The general characteristics of three Dysphania chloroplast genomes
|
Characteristics
| D. ambrosioides | D. pumilio | D. botrys |
|
Accession Number
|
NC_041201
|
NC_041159
|
NC_042166
|
|
References
|
This study
|
(Kim et al., 2019)
|
(Chen and Yang, 2018)
|
|
Total cpDNA size (bp) / GC content (%)
|
151,689 / 36.9
|
151,962 / 36.9
|
152,055 / 36.8
|
|
LSC size (bp) / GC content (%)
|
83,241 / 34.9
|
83,756 / 34.8
|
83,769 / 34.7
|
|
IR size (bp) / GC content (%)
|
25,103 / 42.7
|
25,231 / 42.7
|
25,185 / 42.7
|
|
SSC size (bp) / GC content (%)
|
18,062 / 30.3
|
17,742 / 30.4
|
17,916 / 30.1
|
|
Number of genes
|
128
|
128
|
128
|
|
Number of protein-coding genes
|
84
|
84
|
83
|
|
Number of tRNA genes
|
36
|
36
|
37
|
|
Number of rRNA genes
|
8
|
8
|
8
|
|
Number of duplicated genes
|
17
|
17
|
17
|
Simple sequence repeats analyses of Dysphania chloroplast genomes
In total, 512 simple sequence repeats (SSRs; Table 2 and Appendix 1) including potential (411; 80.47%) and extended SSRs (33; 6.45%) are identified from D. ambrosioides chloroplast genome. 50, 6, and 11 SSRs are identified in LSC, IR, and SSC regions, respectively (Fig. 2), which is larger than those of Chenopodium album and C. quinoa (Hong et al., 2017), and similar to those of Rosamultiflora, R. maximowicziana, and R. luciae chloroplast genomes (Jeon and Kim, 2019). 240, 130, and 42 potential SSRs are also found in LSC, IR, and SSC regions, respectively (Fig. 2), presenting higher density in IR region than that of SSRs. 18, 10, and 5 extended SSRs are in LSC, IR, and SSC regions, showing number of extended SSRs in one IR region is same to that of SSC (Fig. 2). Considering two Dysphania chloroplast genomes, total number of SSRs are quite different: D. pumilio contains 76 SSRs and D. botrys has 45 SSRs. In addition, Chenopodium quinoa and C. album displayed 44 and 53 SSRs, respectively (Hong et al., 2017), reflecting various number of SSRs in both neighbor genera. However, total number of SSRs, potential SSRs, and extended SSRs are similar to each other: D. ambrosioides contains 512, D. pumilio has 509, and D. botrys covers 508 (Fig 2). Based on coordination of SSRs, potential SSRs, and extended SSRs of three Dysphania species, there are many species-specific areas where SSRs in one species not showing corresponding SSRs in the other two species are located, indicating that similarity of total numbers of SSRs, potential SSRs and extended SSRs of three Dysphania species is coincidence.
Table 2. List of simple sequence repeats including two types identified from Dysphania ambrosioides
|
Name
|
Type
|
SSR Type
|
Start
|
End
|
Unit sequence
|
Repeat number
|
Gene
|
|
c70000001
|
ExtendedSSR
|
HeptaSSR
|
1825
|
1838
|
CCAAATA
|
2
| matK
|
|
c70000002
|
ExtendedSSR
|
HeptaSSR
|
3600
|
3613
|
CTCTTAA
|
2
| |
|
c70000003
|
ExtendedSSR
|
HeptaSSR
|
5216
|
5229
|
ACAATTA
|
2
| |
|
cM0000001
|
SSR
|
MonoSSR
|
6699
|
6708
|
A
|
10
| |
|
cM0000002
|
SSR
|
MonoSSR
|
7362
|
7371
|
A
|
10
| |
|
cM0000003
|
SSR
|
MonoSSR
|
7626
|
7637
|
A
|
12
| |
|
cT0000001
|
SSR
|
TriSSR
|
7626
|
7637
|
AAA
|
4
| |
|
cD0000001
|
SSR
|
DiSSR
|
7977
|
7986
|
AT
|
5
| |
|
cM0000004
|
SSR
|
MonoSSR
|
8860
|
8869
|
T
|
10
| |
|
cM0000005
|
SSR
|
MonoSSR
|
9509
|
9518
|
T
|
10
| |
|
cTe0000001
|
SSR
|
TetraSSR
|
12734
|
12745
|
GGAA
|
3
| |
|
cD0000002
|
SSR
|
DiSSR
|
13175
|
13184
|
CA
|
5
| |
|
c80000001
|
ExtendedSSR
|
OctaSSR
|
13777
|
13792
|
TATTTATA
|
2
| |
|
cTe0000002
|
SSR
|
TetraSSR
|
15718
|
15729
|
ATTT
|
3
| |
|
c90000002
|
ExtendedSSR
|
NonaSSR
|
15759
|
15776
|
AAAAGATAA
|
2
| |
|
cM0000006
|
SSR
|
MonoSSR
|
17785
|
17795
|
T
|
11
| rpoC2
|
|
cM0000007
|
SSR
|
MonoSSR
|
21963
|
21972
|
A
|
10
| |
|
cM0000008
|
SSR
|
MonoSSR
|
22174
|
22183
|
A
|
10
| |
|
cM0000009
|
SSR
|
MonoSSR
|
25437
|
25446
|
T
|
10
| rpoB
|
|
c80000002
|
ExtendedSSR
|
OctaSSR
|
26216
|
26231
|
ATATATGA
|
2
| |
|
cM0000010
|
SSR
|
MonoSSR
|
28517
|
28526
|
A
|
10
| |
|
cM0000011
|
SSR
|
MonoSSR
|
30306
|
30315
|
A
|
10
| |
|
cTe0000003
|
SSR
|
TetraSSR
|
31338
|
31349
|
TCTT
|
3
| |
|
cM0000012
|
SSR
|
MonoSSR
|
34378
|
34387
|
T
|
10
| |
|
c80000003
|
ExtendedSSR
|
OctaSSR
|
35168
|
35183
|
CAATATAA
|
2
| |
|
c70000007
|
ExtendedSSR
|
HeptaSSR
|
41592
|
41605
|
TAACAAA
|
2
| |
|
cM0000013
|
SSR
|
MonoSSR
|
42109
|
42118
|
A
|
10
| |
|
c70000008
|
ExtendedSSR
|
HeptaSSR
|
44091
|
44104
|
TTAGTTA
|
2
| |
|
cD0000003
|
SSR
|
DiSSR
|
45286
|
45297
|
TA
|
6
| |
|
c70000009
|
ExtendedSSR
|
HeptaSSR
|
45333
|
45346
|
TAAATGA
|
2
| |
|
cD0000004
|
SSR
|
DiSSR
|
45636
|
45649
|
AT
|
7
| |
|
cM0000014
|
SSR
|
MonoSSR
|
48064
|
48077
|
T
|
14
| |
|
cT0000002
|
SSR
|
TriSSR
|
48064
|
48075
|
TTT
|
4
| |
|
cM0000015
|
SSR
|
MonoSSR
|
50198
|
50210
|
A
|
13
| |
|
cT0000003
|
SSR
|
TriSSR
|
50198
|
50209
|
AAA
|
4
| |
|
c70000011
|
ExtendedSSR
|
HeptaSSR
|
50233
|
50246
|
TTTGTTA
|
2
| |
|
cM0000016
|
SSR
|
MonoSSR
|
50602
|
50613
|
T
|
12
| |
|
cT0000004
|
SSR
|
TriSSR
|
50602
|
50613
|
TTT
|
4
| |
|
cM0000017
|
SSR
|
MonoSSR
|
50825
|
50835
|
T
|
11
| |
|
cM0000018
|
SSR
|
MonoSSR
|
53712
|
53721
|
T
|
10
| atpB
|
|
cM0000019
|
SSR
|
MonoSSR
|
53827
|
53836
|
A
|
10
| |
|
cM0000020
|
SSR
|
MonoSSR
|
54092
|
54101
|
T
|
10
| |
|
c70000012
|
ExtendedSSR
|
HeptaSSR
|
56336
|
56349
|
TCTTATA
|
2
| |
|
c80000004
|
ExtendedSSR
|
OctaSSR
|
56405
|
56420
|
TTTCTTTT
|
2
| |
|
c70000013
|
ExtendedSSR
|
HeptaSSR
|
58357
|
58370
|
AATTCGT
|
2
| |
|
cM0000021
|
SSR
|
MonoSSR
|
58435
|
58444
|
T
|
10
| |
|
cTe0000004
|
SSR
|
TetraSSR
|
58516
|
58531
|
TAAT
|
4
| |
|
c80000006
|
ExtendedSSR
|
OctaSSR
|
58677
|
58692
|
TTCTTTAT
|
2
| psaI
|
|
cM0000022
|
SSR
|
MonoSSR
|
60250
|
60259
|
A
|
10
| cemA
|
|
cTe0000005
|
SSR
|
TetraSSR
|
60938
|
60949
|
TGAA
|
3
| cemA
|
|
cM0000023
|
SSR
|
MonoSSR
|
64357
|
64367
|
A
|
11
| |
|
cD0000005
|
SSR
|
DiSSR
|
64561
|
64572
|
AT
|
6
| |
|
cM0000024
|
SSR
|
MonoSSR
|
64745
|
64755
|
T
|
11
| |
|
cM0000025
|
SSR
|
MonoSSR
|
65181
|
65191
|
T
|
11
| |
|
cP0000001
|
SSR
|
PentaSSR
|
65399
|
65413
|
TTTAT
|
3
| |
|
cM0000026
|
SSR
|
MonoSSR
|
66109
|
66118
|
T
|
10
| |
|
cM0000027
|
SSR
|
MonoSSR
|
67593
|
67602
|
T
|
10
| |
|
cM0000028
|
SSR
|
MonoSSR
|
68229
|
68238
|
T
|
10
| |
|
cM0000029
|
SSR
|
MonoSSR
|
68241
|
68251
|
T
|
11
| |
|
cM0000030
|
SSR
|
MonoSSR
|
68759
|
68769
|
T
|
11
| |
|
cM0000031
|
SSR
|
MonoSSR
|
70683
|
70692
|
T
|
10
| |
|
c70000015
|
ExtendedSSR
|
HeptaSSR
|
71719
|
71732
|
CTGGTTG
|
2
| psbB
|
|
cM0000032
|
SSR
|
MonoSSR
|
77245
|
77254
|
T
|
10
| rpoA
|
|
c70000016
|
ExtendedSSR
|
HeptaSSR
|
78650
|
78663
|
TTTTAGT
|
2
| |
|
cM0000033
|
SSR
|
MonoSSR
|
80264
|
80273
|
A
|
10
| |
|
c70000017
|
ExtendedSSR
|
HeptaSSR
|
80951
|
80964
|
TAAATAT
|
2
| |
|
cM0000034
|
SSR
|
MonoSSR
|
81501
|
81510
|
T
|
10
| |
|
cM0000035
|
SSR
|
MonoSSR
|
83182
|
83197
|
T
|
16
| rpl22
|
|
cT0000005
|
SSR
|
TriSSR
|
83182
|
83196
|
TTT
|
5
| rpl22
|
|
c90000004
|
ExtendedSSR
|
NonaSSR
|
87117
|
87134
|
GGAACATTT
|
2
| ycf2
|
|
cD0000006
|
SSR
|
DiSSR
|
92466
|
92475
|
TA
|
5
| |
|
cM0000036
|
SSR
|
MonoSSR
|
97844
|
97853
|
T
|
10
| |
|
c80000008
|
ExtendedSSR
|
OctaSSR
|
102025
|
102040
|
TTTTGAGA
|
2
| |
|
cTe0000006
|
SSR
|
TetraSSR
|
104017
|
104028
|
AGGT
|
3
| rrn23
|
|
c70000019
|
ExtendedSSR
|
HeptaSSR
|
106379
|
106392
|
TATGTTT
|
2
| |
|
c70000020
|
ExtendedSSR
|
HeptaSSR
|
106658
|
106671
|
AAGAATG
|
2
| |
|
c90000005
|
ExtendedSSR
|
NonaSSR
|
107219
|
107236
|
GAAGAAGGA
|
2
| ycf1
|
|
cM0000037
|
SSR
|
MonoSSR
|
108408
|
108417
|
A
|
10
| ycf1
|
|
c70000021
|
ExtendedSSR
|
HeptaSSR
|
110105
|
110118
|
CGAAACT
|
2
| ndhF
|
|
c90000006
|
ExtendedSSR
|
NonaSSR
|
111025
|
111042
|
AAAGTCAAT
|
2
| |
|
cM0000038
|
SSR
|
MonoSSR
|
111305
|
111315
|
A
|
11
| |
|
cD0000007
|
SSR
|
DiSSR
|
111447
|
111456
|
TA
|
5
| |
|
cD0000008
|
SSR
|
DiSSR
|
112099
|
112108
|
TA
|
5
| |
|
cM0000039
|
SSR
|
MonoSSR
|
112430
|
112439
|
T
|
10
| |
|
c100000001
|
ExtendedSSR
|
DecaSSR
|
112770
|
112789
|
ATATATAGTT
|
2
| |
|
cM0000040
|
SSR
|
MonoSSR
|
112866
|
112876
|
A
|
11
| |
|
cM0000041
|
SSR
|
MonoSSR
|
113133
|
113142
|
T
|
10
| |
|
c70000023
|
ExtendedSSR
|
HeptaSSR
|
117365
|
117378
|
TAGAATA
|
2
| ndhG
|
|
c70000024
|
ExtendedSSR
|
HeptaSSR
|
118129
|
118142
|
ATTTCCA
|
2
| ndhI
|
|
cM0000042
|
SSR
|
MonoSSR
|
118476
|
118486
|
T
|
11
| |
|
cT0000006
|
SSR
|
TriSSR
|
120629
|
120640
|
TGT
|
4
| ndhA
|
|
cTe0000007
|
SSR
|
TetraSSR
|
122441
|
122452
|
TATT
|
3
| |
|
cTe0000008
|
SSR
|
TetraSSR
|
124069
|
124080
|
TAAT
|
3
| ycf1
|
|
cM0000043
|
SSR
|
MonoSSR
|
126694
|
126703
|
T
|
10
| ycf1
|
|
c90000007
|
ExtendedSSR
|
NonaSSR
|
127875
|
127892
|
TCCTTCTTC
|
2
| ycf1
|
|
c70000025
|
ExtendedSSR
|
HeptaSSR
|
128438
|
128451
|
TTCATTC
|
2
| |
|
c70000026
|
ExtendedSSR
|
HeptaSSR
|
128719
|
128732
|
AAACATA
|
2
| |
|
cTe0000009
|
SSR
|
TetraSSR
|
131081
|
131092
|
CTAC
|
3
| rrn23
|
|
c80000009
|
ExtendedSSR
|
OctaSSR
|
133071
|
133086
|
TCTCAAAA
|
2
| |
|
cM0000044
|
SSR
|
MonoSSR
|
137258
|
137267
|
A
|
10
| |
|
cD0000009
|
SSR
|
DiSSR
|
142635
|
142644
|
AT
|
5
| |
|
c90000008
|
ExtendedSSR
|
NonaSSR
|
147977
|
147994
|
AAATGTTCC
|
2
| ycf2
|
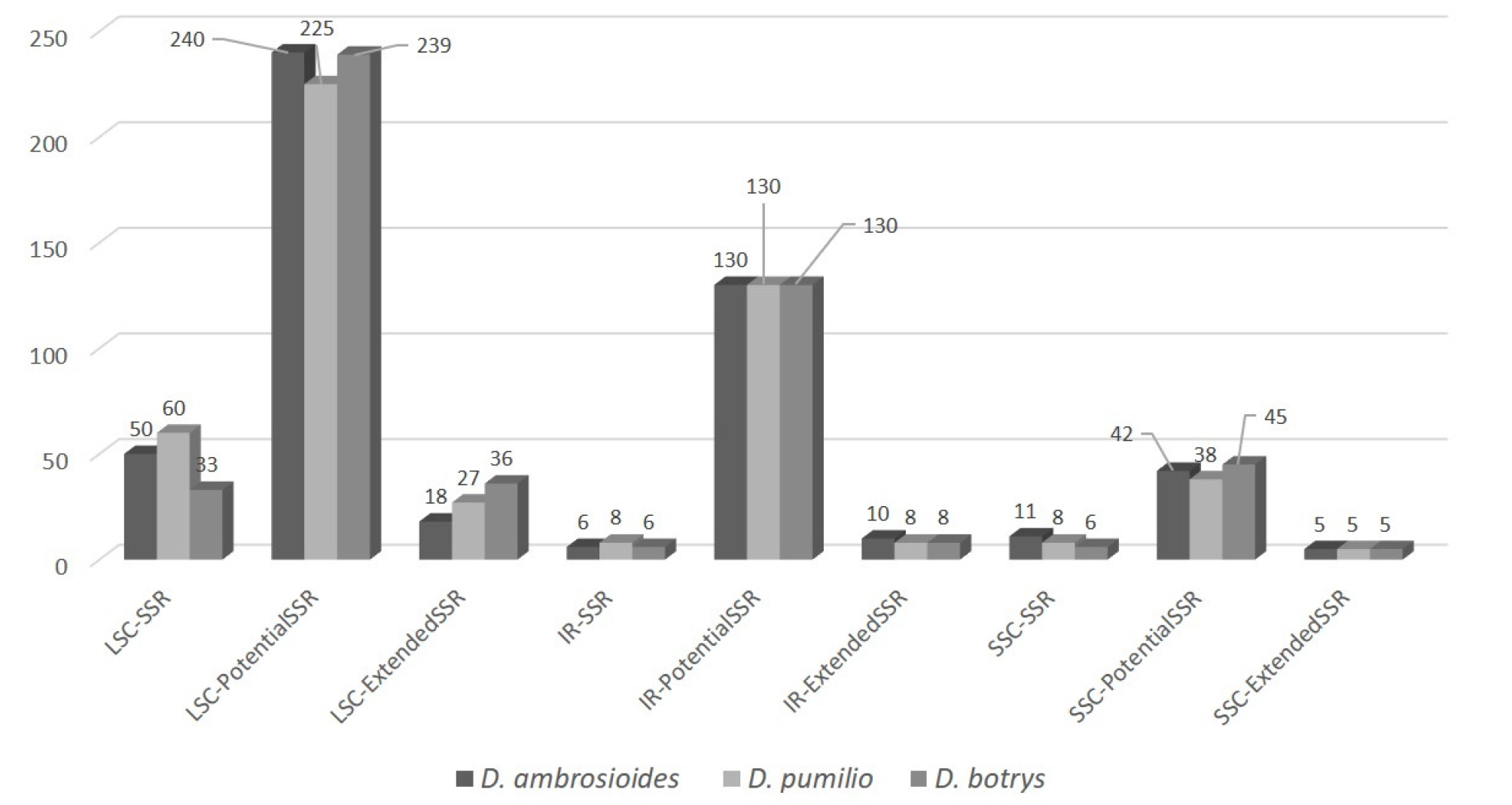
Fig. 2.
Simple sequence repeat sequences of three Dysphania species along with three regions, LSC, IR, and SSC. X-axis presents three regions of chloroplast genomes, LSC, IR, and SSC along with three SSR types, SSRs, Potential SSRs, and Extended SSRs. Y-axis indicates number of SSRs. Three species were presented with different grey colors.
Along with types of SSRs (MonoSSR to HexaSSR), D. ambrosioides contains no PentaSSR and HexaSSR; while the other two Dysphania species have one PentaSSR (Fig 3). MonoSSR shows the highest number of SSRs among the six SSR types for all three Dysphania species, which is same to those of C. quinoa, C. album (Hong et al., 2017), Haloxylon ammodendron and H. persicum (Dong et al., 2016) in Chenopodiaceae and the three Rosa species in Rosaceae (Jeon and Kim, 2019; Kim et al., 2019; Li et al., 2019). However, trend of numbers of DiSSRs, TriSSRs, and TetraSSRs presents difference among three species: D. ambrosioides presents that number of DiSSRs is same to that of TetraSSRs and number of TriSSRs is the lowest, same trend to D. botrys (Fig 3). D. pumilio shows that number of DiSSRs is same to that of TetraSSRs, same to D. ambrosioides, but number of TriSSRs is higher than those of DiSSRs and TetraSSRs (Fig 3). Potential SSRs of which repeat number is 2 displays the same trend of PentaSSRs and HexaSSRs among three species and that of PentaSSRs is larger than that of HexaSSRs. Extended SSRs ranging from HeptaSSR to DecaSSR also present the same trend among three Dysphania species. Development of molecular markers for distinguishing Dysphania species as well as their populations, these differences can be utilized for better resolutions (Song et al., 2003; Wang et al., 2009; Würschum et al., 2013).
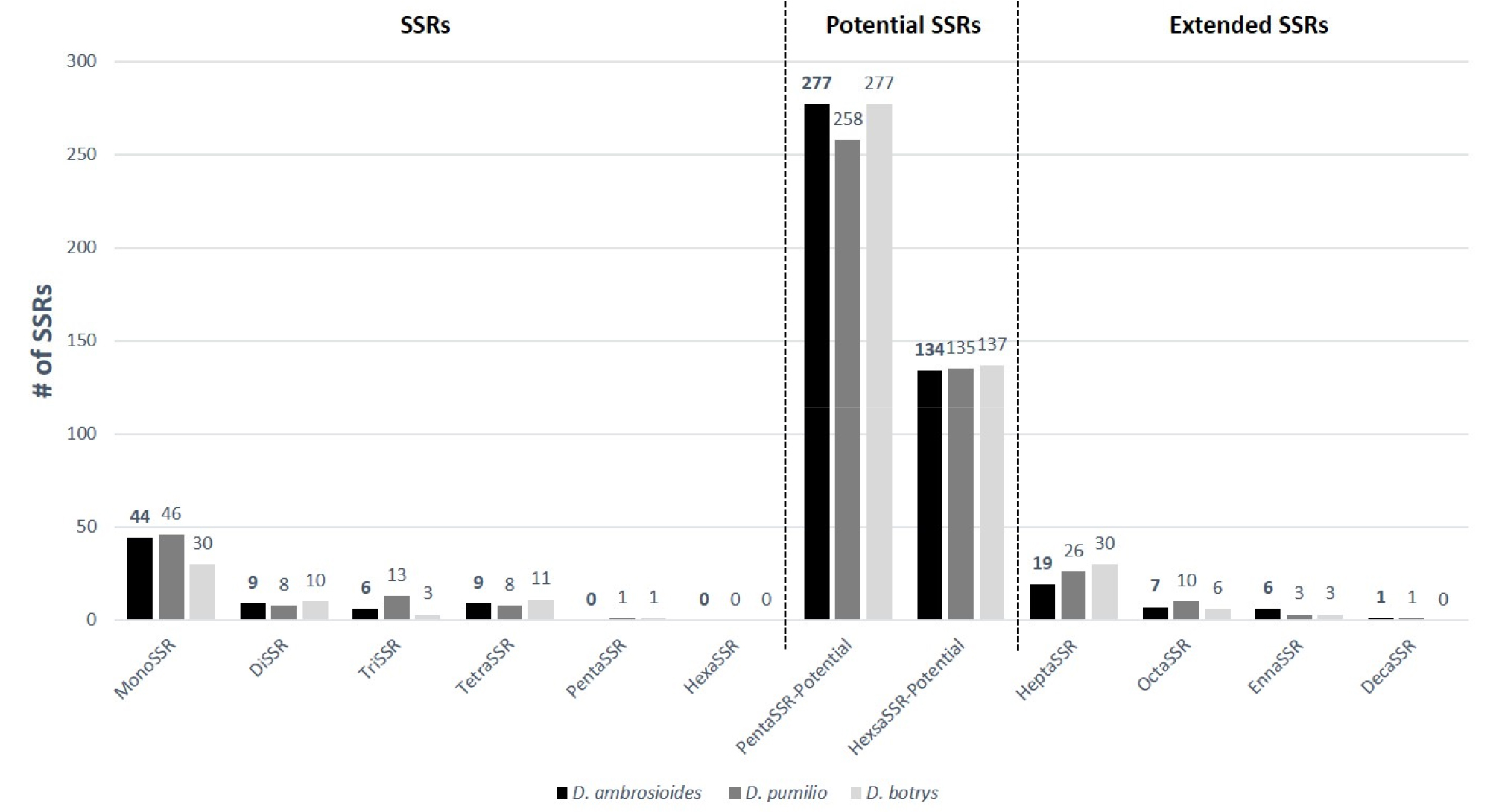
Fig. 3.
Simple sequence repeat sequences of three Dysphania species along with their unit length. X-axis presents types of SSRs of which length is from 1 bp (MonoSSR) to 10 bp (DecaSSR) classified by three SSRs displayed on the top of the graph: SSRs, Potential SSRs, and Extended SSRs. Y-axis indicates number of SSRs. Dark black, grey, and light grey bars represent SSRs of D. ambrosioides, D. pumilio, and D. botrys.
14 out of 68 SSRs are found in nine genes, rpoC2, rpoB, atpB, cemA, rpoA, rpl22, ycf1, ndhA as well as rrn23, ribosomal RNA, consisting of eight MonoSSRs, four TetraSSRs, and two TriSSRs (Table 2). 4 out of 33 extended SSRs covers four genes, ycf2, trnV-GAC, psbA, and rpoC2, which are different from eight genes containing SSRs (Table 2). Potential SSRs also covers many genes of which some are overlapped with genes containging SSRs (Appendix 1). These SSRs identified in protein-coding genes will be utilized for better population genetics studies because it may link to essential functions inside cell.
Nucleotide diversity analysis within three Dysphania species
The complete chloroplast sequences of Dysphania ambrosioides, D. botrys, and D. pumilio were aligned with MAFFT 7.388 (Katoh and Standley, 2013; Fig. 4). Overall nucleotide diversity (Pi) was 0.0068 (Fig 4). Based on nucleotide diversity across three chloroplast genome of Dysphania species, IR region presents very low nucleotide diversity and LSC and SSC regions present higher nucleotide diversity than that of IR as same as the other chloroplast genomes (Hong et al., 2017; Thomson et al., 2017; Li et al., 2018).
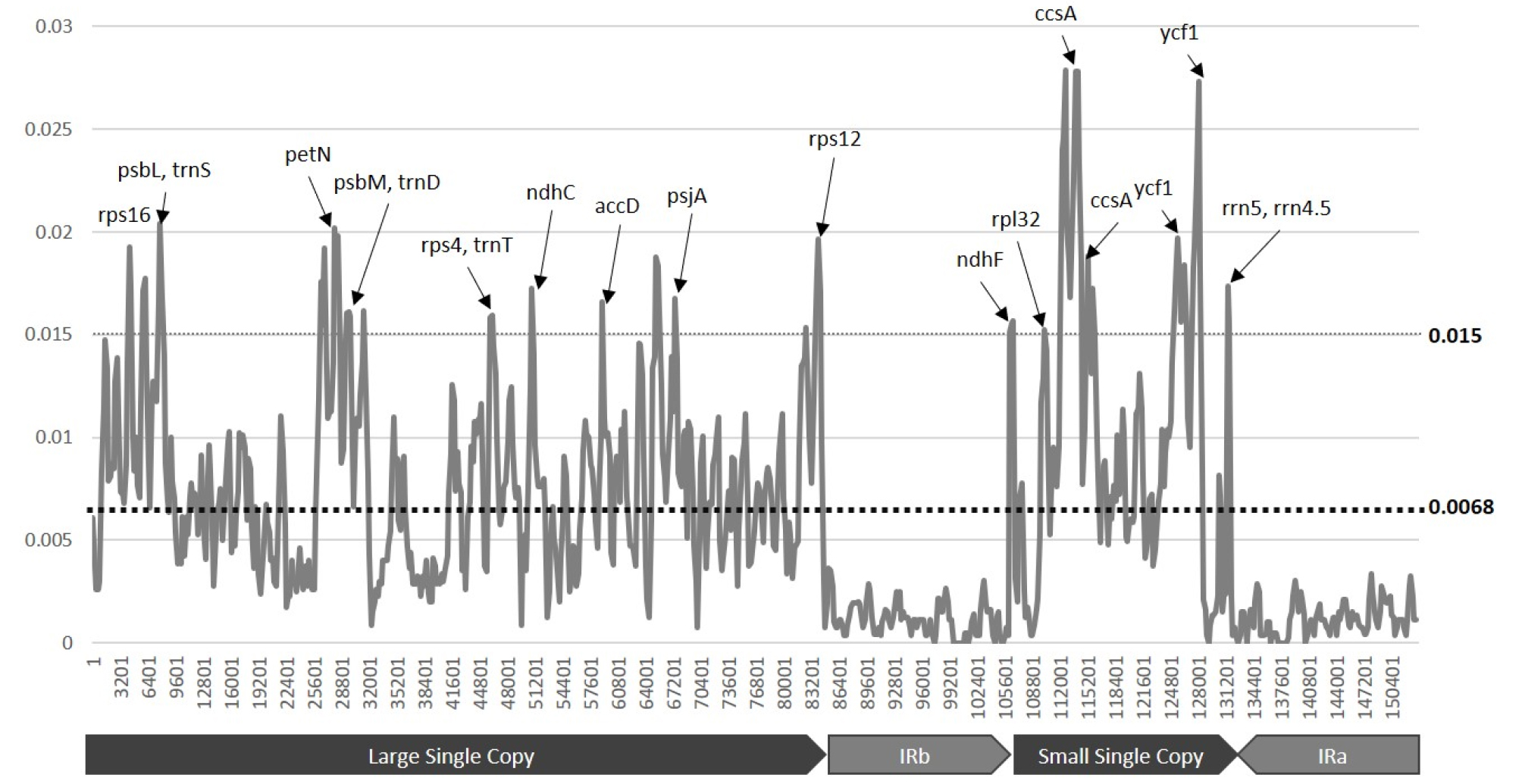
Fig. 4.
Nucleotide diversity of three Dysphania chloroplast genomes. X-axis presents chloroplast genomic coordination and Y-axis shows nucleotide diversity value (Pi) calculated with 500-bp windows and 200-bp step. Black arrows and gene names present nucleotide diversity peak of which value is above 0.015. Below X-axis, arrow diagrams show four regions, LSC, IRb, SSC, and IRa, respectively.
Based on nucleotide diversity distribution along with Dysphania chloroplast genomes (Fig. 4), rps16, psbL, trnS, petN, psbM, trnD, rps4, trnT, ndhC, accD, psbA, rps12, ndhF, rpl32, ccsA, ycf1, rrn5 and rrn4.5 genes present high nucleotide diversity (> 0.015), which are candidates for molecular markers of population genetics in Dysphania species. Interesting thing is that some part of ribosomal RNAs, such as rrn5 and rrn4.5, present high nucleotide diversity value, which is same trend to that of Chenopodium (Hong et al., 2017); while different from those of Rosa and Symplocarpus species (Jeon and Kim, 2019; Kim et al., 2019). Number of Dysphania genes with high nucleotide diversity are larger than those of Chenopodium (Hong et al., 2017) and Rosa species (Jeon and Kim, 2019); while similar to that of Symplocarpus species (Kim et al., 2019) indicating that this is species or genus-specific feature.
Comparative analysis of IR junction structure
IR region on chloroplast genome is one of main sources to expand or to shrink chloroplast genome sequence (Asaf et al., 2016; Dong et al., 2016; Kim et al., 2019; Li et al., 2018; Yang et al., 2016). Based on alignment of three Dysphania chloroplast genomes, 3’ end position of ndhF at the border of IR and SSC are different; D. ambrosioides present the shortest ndhF gene; while ycf1 in the end of IR is ended in the border for three chloroplast genomes (Fig. 5). It is similar to the cases of i) some bamboo species having pseudo-ndhF genes because of different IR borders (Wang et al., 2018) and ii) different size of ndhF genes located in the start part of SSC across Solanaceae species (Chung et al., 2006). Stop codons of ycf1 genes of three Dysphania species are located in the same position, the end of IR (Fig. 5), which is different from the comparison results: some of ycf1 genes are extended to SSC region (Chung et al., 2006; Hong et al., 2017; Xie et al., 2018). There is no difference among three Dysphania chloroplast genomes in two junctions between SSC and IRa and IRa and LSC (Fig. 5); while lengths of SSC of three Dysphania present that the longest is D.ambrosioides, the second is D. botrys, and the shortest is D. pumilio. This indicates that common ancestor of D. pumilio and D. botrys may have shorter SSC than that of D. ambrosioides based on phylogenetic relationship (Fig. 6).
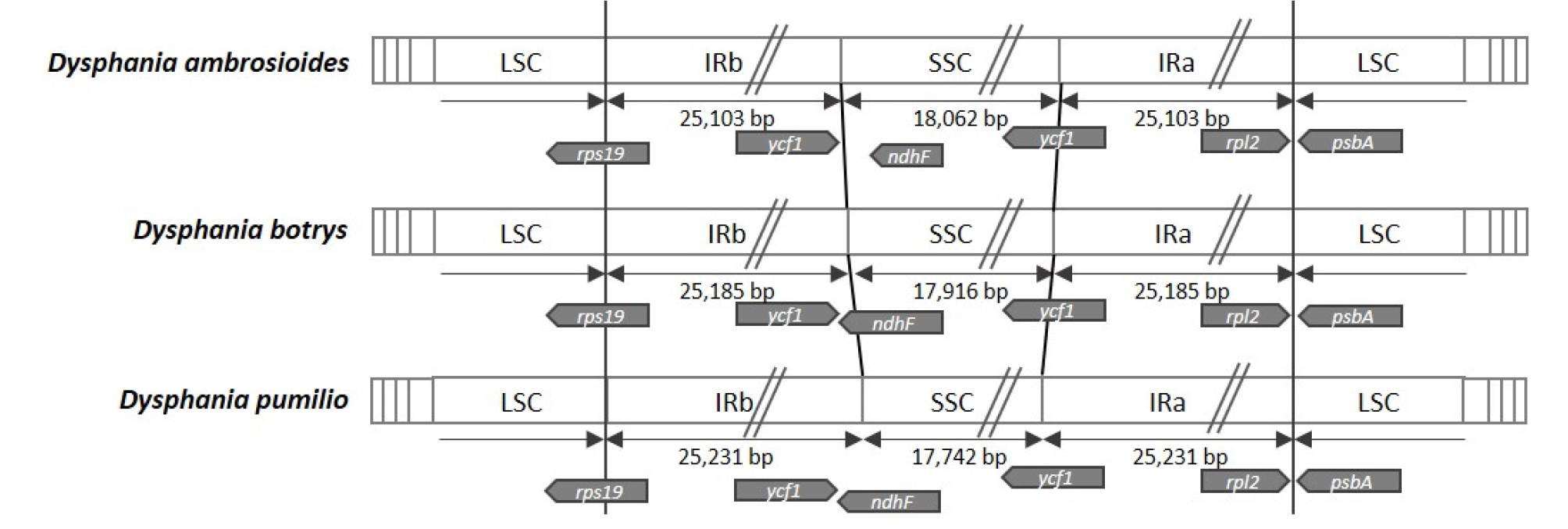
Fig. 5.
Comparison of the border positions of IR, SSC, and LSC regions among three Dysphania species. Diagrams present three chloroplast genomes of Dysphania with each region. Black arrows show length of each region except LSC, and gray arrow diagrams show genes located in junctions between LSC and IRb, IRb and SSC, SSC and IRa, and IRa and LSC.
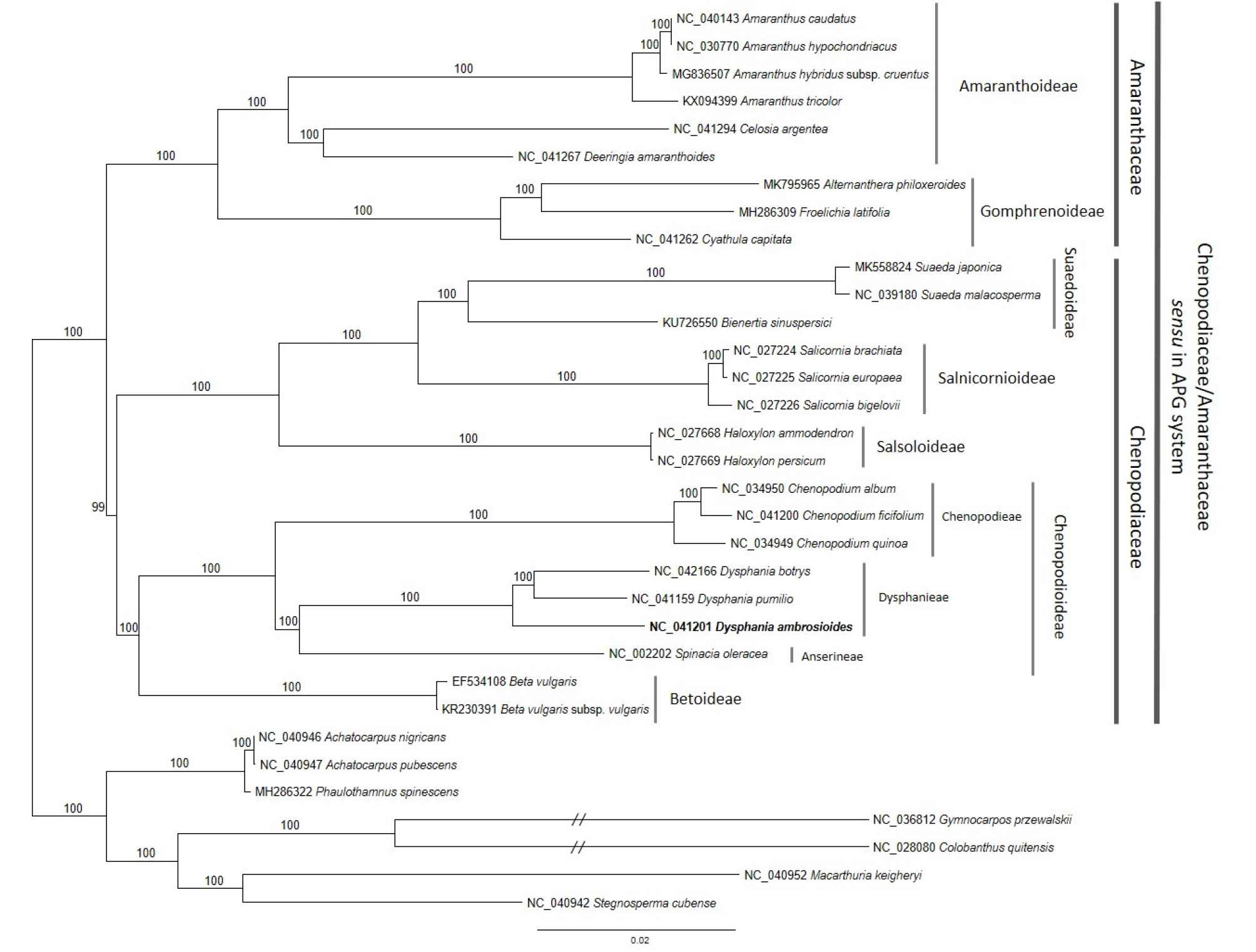
Fig. 6.
Maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 26 Chenopodiaceae/Amaranthaceae sensu in APG complete chloroplast genomes. The numbers above branches indicate bootstrap support values of neighbor joining, maximum likelihood, and minimum evolution phylogenetic trees. Names on gray bars present tribes and subfamilies (light gray) and families (dark gray). Bolded species name indicates our chloroplast genome.
Phylogenetic analyses with complete chloroplast genomes
A total of 186,232 aligned nucleotide bases included 129,316 constant sites (47.6%) and 37,043 parsimony-informative sites (19.9%) were identified from multiple sequence alignments of 26 chloroplast genomes. The bootstrapped ML tree presents that genus Dysphania strongly supported monophyly as previous studies displayed (Fuentes-Bazan et al., 2012a; Fuentes-Bazan et al., 2012b; Kadereit et al., 2003; Kadereit et al., 2010) and formed sister clade with Spinacia (Fig. 6). D. ambrosioides has basal position in Dysphania clade with clustering the rest of two Dysphania species, D. pumilio and D. botrys (Fig. 6). In the level of tribe, two tribes in Chenopodioidae, Dysphanieae including genus Dysphania and Anserineae containing Spinacia oleracea have shown week or no resolution on their phylogenetic relation (Fuentes-Bazan et al., 2012b; Kadereit et al., 2003; Kadereit et al., 2010; Sukhorukov et al., 2018). Although the number of taxa is less than those in previous studies using molecular markers, complete chloroplast genome sequences provide much higher resolution of phylogenetic tree due to large amount of sequences; this unclear relationship is clearly resolved in our phylogenetic tree (Fig. 6). In addition, recent phylogenetic trees based on complete chloroplast genome sequences also presented the same topology (Chen and Yang, 2018; Kim et al., 2019; Park and Kim, 2019), supporting phylogenetic relationship between Dysphanieae and Anserineae becomes clear.
Conclusion
We determined the complete chloroplast sequence of useful medicinal and invasive species Dysphania ambrosioides and compared this species with other chloroplast genome of two Dysphania species. Chloroplast genome of D. ambrosioides presents 151,689 bp long with four subregions: 83,421 bp of large single copy and 18,062 bp of small single copy regions separated by 25,103 bp of inverted repeat regions and its GC ratio is 36.9%, presenting conserved manner among three Dysphania chloroplast genomes. SSR analysis presents that common features as well as differences among three Dysphania chloroplast genomes can be utilized for further population genetic researches. Nucleotide diversity of Dysphania chloroplast genomes 18 genes including two ribosomal RNAs contains high nucleotide diversity peaks, which may be genus or species-specific manner. The phylogenetic position of genus Dysphania was supported as in the previous studies, and D. ambrosioides was found to be the most basal in Dysphania among three species sequenced so far. The both tribes Dysphanieae and Anserineae were demonstrated in its phylogenetic relationship as sister. Taken together, our investigations of D. ambrosioides chloroplast genomes will be used for further in-depth study of genus Dysphania.
Appendix 1. List of simple sequence repeats of potential SSR identified from
Dysphania ambrosioides|
Name
|
Type
|
SSR Type
|
Start
|
End
|
Unit sequence
|
Repeat number
|
Gene
|
|
cH0000001
|
PotentialSSR
|
PentaSSR
|
197
|
206
|
TAAAA
|
2
| |
|
cP0000002
|
PotentialSSR
|
HexaSSR
|
369
|
380
|
TTGAAA
|
2
| psbA
|
|
cP0000003
|
PotentialSSR
|
PentaSSR
|
1293
|
1302
|
ATTTA
|
2
| |
|
cH0000002
|
PotentialSSR
|
PentaSSR
|
1759
|
1768
|
CATTT
|
2
| |
|
cH0000003
|
PotentialSSR
|
HexaSSR
|
2628
|
2639
|
AGAAAA
|
2
| matK
|
|
cH0000004
|
PotentialSSR
|
HexaSSR
|
3045
|
3056
|
TTGTGC
|
2
| matK
|
|
cP0000004
|
PotentialSSR
|
HexaSSR
|
3303
|
3314
|
ATTTCA
|
2
| matK
|
|
cP0000005
|
PotentialSSR
|
PentaSSR
|
4085
|
4094
|
TATGT
|
2
| |
|
cP0000006
|
PotentialSSR
|
PentaSSR
|
4270
|
4279
|
CATTT
|
2
| |
|
cP0000007
|
PotentialSSR
|
PentaSSR
|
4280
|
4289
|
TTTTA
|
2
| |
|
cP0000008
|
PotentialSSR
|
PentaSSR
|
4519
|
4528
|
AAAAT
|
2
| |
|
cP0000009
|
PotentialSSR
|
PentaSSR
|
4576
|
4585
|
AGGAA
|
2
| |
|
cP0000010
|
PotentialSSR
|
PentaSSR
|
4871
|
4880
|
TAGAT
|
2
| |
|
cP0000011
|
PotentialSSR
|
PentaSSR
|
4946
|
4955
|
AAAGT
|
2
| |
|
cP0000012
|
PotentialSSR
|
PentaSSR
|
5029
|
5038
|
TTTCA
|
2
| |
|
cP0000013
|
PotentialSSR
|
PentaSSR
|
5106
|
5115
|
AAAAG
|
2
| |
|
cP0000014
|
PotentialSSR
|
PentaSSR
|
5126
|
5135
|
CATTT
|
2
| |
|
cH0000005
|
PotentialSSR
|
PentaSSR
|
5359
|
5368
|
AAAGA
|
2
| |
|
cP0000015
|
PotentialSSR
|
HexaSSR
|
5535
|
5546
|
ACCCTA
|
2
| |
|
cP0000016
|
PotentialSSR
|
PentaSSR
|
5551
|
5560
|
CTTCT
|
2
| |
|
cP0000017
|
PotentialSSR
|
PentaSSR
|
5654
|
5663
|
GAACA
|
2
| |
|
cH0000006
|
PotentialSSR
|
PentaSSR
|
5798
|
5807
|
AATTT
|
2
| |
|
cP0000018
|
PotentialSSR
|
HexaSSR
|
5968
|
5979
|
AGAAAT
|
2
| |
|
cP0000019
|
PotentialSSR
|
PentaSSR
|
6467
|
6476
|
TTCTA
|
2
| |
|
cP0000020
|
PotentialSSR
|
PentaSSR
|
6518
|
6527
|
CCAAA
|
2
| |
|
cH0000007
|
PotentialSSR
|
PentaSSR
|
7202
|
7211
|
TAATA
|
2
| |
|
cP0000021
|
PotentialSSR
|
HexaSSR
|
7243
|
7254
|
TTTCTT
|
2
| |
|
cH0000008
|
PotentialSSR
|
PentaSSR
|
7306
|
7315
|
TCAAA
|
2
| |
|
cP0000022
|
PotentialSSR
|
HexaSSR
|
7674
|
7685
|
AAATAG
|
2
| |
|
cP0000025
|
PotentialSSR
|
PentaSSR
|
8150
|
8159
|
AAAGA
|
2
| |
|
cH0000009
|
PotentialSSR
|
PentaSSR
|
8226
|
8235
|
CAGGC
|
2
| |
|
cH0000010
|
PotentialSSR
|
HexaSSR
|
8286
|
8297
|
TGATAA
|
2
| |
|
cP0000026
|
PotentialSSR
|
HexaSSR
|
8314
|
8325
|
AAAGCA
|
2
| |
|
cP0000027
|
PotentialSSR
|
PentaSSR
|
8344
|
8353
|
AACAT
|
2
| |
|
cH0000011
|
PotentialSSR
|
PentaSSR
|
8957
|
8966
|
AAACA
|
2
| |
|
cP0000028
|
PotentialSSR
|
HexaSSR
|
8987
|
8998
|
AAATAG
|
2
| |
|
cP0000029
|
PotentialSSR
|
PentaSSR
|
9068
|
9077
|
TTGAA
|
2
| |
|
cP0000030
|
PotentialSSR
|
PentaSSR
|
9357
|
9366
|
TCTCA
|
2
| |
|
cH0000012
|
PotentialSSR
|
PentaSSR
|
9388
|
9397
|
CAAAA
|
2
| |
|
cP0000031
|
PotentialSSR
|
HexaSSR
|
10312
|
10323
|
GCTTGT
|
2
| atpA
|
|
cH0000013
|
PotentialSSR
|
PentaSSR
|
11746
|
11755
|
TCTCT
|
2
| |
|
cP0000032
|
PotentialSSR
|
HexaSSR
|
11977
|
11988
|
CAATAA
|
2
| |
|
cP0000033
|
PotentialSSR
|
PentaSSR
|
12580
|
12589
|
TAAAT
|
2
| |
|
cH0000014
|
PotentialSSR
|
PentaSSR
|
12630
|
12639
|
ACTTA
|
2
| |
|
cP0000034
|
PotentialSSR
|
HexaSSR
|
12901
|
12912
|
CTTTTC
|
2
| |
|
cP0000035
|
PotentialSSR
|
PentaSSR
|
13448
|
13457
|
ATTCA
|
2
| |
|
cP0000036
|
PotentialSSR
|
PentaSSR
|
13599
|
13608
|
CTATT
|
2
| |
|
cH0000016
|
PotentialSSR
|
PentaSSR
|
13799
|
13808
|
AAATA
|
2
| |
|
cH0000017
|
PotentialSSR
|
HexaSSR
|
13852
|
13863
|
CATATA
|
2
| |
|
cH0000018
|
PotentialSSR
|
HexaSSR
|
14225
|
14236
|
TAAAGC
|
2
| atpI
|
|
cP0000037
|
PotentialSSR
|
HexaSSR
|
14722
|
14733
|
ATTTAA
|
2
| |
|
cP0000038
|
PotentialSSR
|
PentaSSR
|
15641
|
15650
|
TTTCT
|
2
| |
|
cP0000039
|
PotentialSSR
|
PentaSSR
|
15736
|
15745
|
TAAAT
|
2
| |
|
cH0000019
|
PotentialSSR
|
PentaSSR
|
16792
|
16801
|
CAATT
|
2
| rpoC2
|
|
cP0000040
|
PotentialSSR
|
HexaSSR
|
17022
|
17033
|
AATTGG
|
2
| rpoC2
|
|
cP0000041
|
PotentialSSR
|
PentaSSR
|
17242
|
17251
|
TGATC
|
2
| rpoC2
|
|
cP0000042
|
PotentialSSR
|
PentaSSR
|
17607
|
17616
|
CAAAA
|
2
| rpoC2
|
|
cH0000020
|
PotentialSSR
|
PentaSSR
|
17727
|
17736
|
TATCT
|
2
| rpoC2
|
|
cH0000021
|
PotentialSSR
|
HexaSSR
|
18408
|
18419
|
TTGATC
|
2
| rpoC2
|
|
cP0000043
|
PotentialSSR
|
HexaSSR
|
18780
|
18791
|
ACGTGT
|
2
| rpoC2
|
|
cH0000022
|
PotentialSSR
|
PentaSSR
|
18992
|
19001
|
CATAA
|
2
| rpoC2
|
|
cP0000044
|
PotentialSSR
|
HexaSSR
|
21474
|
21485
|
CAAATC
|
2
| rpoC2
|
|
cP0000045
|
PotentialSSR
|
PentaSSR
|
21588
|
21597
|
GGATT
|
2
| rpoC2
|
|
cH0000023
|
PotentialSSR
|
PentaSSR
|
21794
|
21803
|
CCAAA
|
2
| |
|
cP0000046
|
PotentialSSR
|
HexaSSR
|
21877
|
21888
|
TTTTTA
|
2
| |
|
cH0000024
|
PotentialSSR
|
PentaSSR
|
21892
|
21901
|
AATTA
|
2
| |
|
cH0000025
|
PotentialSSR
|
HexaSSR
|
22061
|
22072
|
TAAAGC
|
2
| |
|
cP0000048
|
PotentialSSR
|
PentaSSR
|
26314
|
26323
|
AAAGG
|
2
| |
|
cP0000049
|
PotentialSSR
|
PentaSSR
|
26488
|
26497
|
TATTG
|
2
| |
|
cH0000026
|
PotentialSSR
|
PentaSSR
|
26522
|
26531
|
TTAAA
|
2
| |
|
cH0000027
|
PotentialSSR
|
HexaSSR
|
26645
|
26656
|
AATTGA
|
2
| |
|
cP0000050
|
PotentialSSR
|
HexaSSR
|
26657
|
26668
|
GAAAAA
|
2
| |
|
cP0000051
|
PotentialSSR
|
PentaSSR
|
26796
|
26805
|
AGTCA
|
2
| |
|
cP0000052
|
PotentialSSR
|
PentaSSR
|
26888
|
26897
|
CAAAA
|
2
| |
|
cH0000028
|
PotentialSSR
|
PentaSSR
|
27019
|
27028
|
GTCTA
|
2
| |
|
cP0000053
|
PotentialSSR
|
HexaSSR
|
27666
|
27677
|
AAAAAG
|
2
| |
|
cH0000029
|
PotentialSSR
|
PentaSSR
|
27740
|
27749
|
AATTT
|
2
| |
|
cP0000055
|
PotentialSSR
|
PentaSSR
|
28151
|
28160
|
TCAAT
|
2
| |
|
cP0000056
|
PotentialSSR
|
PentaSSR
|
28161
|
28170
|
TACTT
|
2
| |
|
cP0000057
|
PotentialSSR
|
PentaSSR
|
28531
|
28540
|
AATCG
|
2
| |
|
cP0000058
|
PotentialSSR
|
PentaSSR
|
28541
|
28550
|
ATAGT
|
2
| |
|
cP0000059
|
PotentialSSR
|
PentaSSR
|
28583
|
28592
|
ATACT
|
2
| |
|
cP0000060
|
PotentialSSR
|
PentaSSR
|
28929
|
28938
|
TATCA
|
2
| |
|
cP0000061
|
PotentialSSR
|
PentaSSR
|
29169
|
29178
|
AAAAG
|
2
| |
|
cP0000062
|
PotentialSSR
|
PentaSSR
|
29565
|
29574
|
ATTTT
|
2
| |
|
cP0000063
|
PotentialSSR
|
PentaSSR
|
29934
|
29943
|
TACCC
|
2
| trnE-UUC
|
|
cP0000065
|
PotentialSSR
|
PentaSSR
|
30745
|
30754
|
ATTAT
|
2
| |
|
cP0000066
|
PotentialSSR
|
PentaSSR
|
30758
|
30767
|
GATAA
|
2
| |
|
cP0000067
|
PotentialSSR
|
PentaSSR
|
30823
|
30832
|
TGGAA
|
2
| |
|
cP0000068
|
PotentialSSR
|
PentaSSR
|
30983
|
30992
|
CAATT
|
2
| |
|
cP0000069
|
PotentialSSR
|
PentaSSR
|
31094
|
31103
|
TTTCT
|
2
| |
|
cP0000070
|
PotentialSSR
|
PentaSSR
|
31251
|
31260
|
TTTCA
|
2
| |
|
cP0000071
|
PotentialSSR
|
PentaSSR
|
31512
|
31521
|
TTAAT
|
2
| |
|
cP0000072
|
PotentialSSR
|
PentaSSR
|
31663
|
31672
|
AAATC
|
2
| |
|
cP0000073
|
PotentialSSR
|
PentaSSR
|
31841
|
31850
|
CGTTT
|
2
| psbD
|
|
cH0000030
|
PotentialSSR
|
PentaSSR
|
32471
|
32480
|
AACCC
|
2
| psbD
|
|
cP0000074
|
PotentialSSR
|
HexaSSR
|
32828
|
32839
|
AACTTT
|
2
| psbC
|
|
cH0000031
|
PotentialSSR
|
PentaSSR
|
33564
|
33573
|
GTCTG
|
2
| psbC
|
|
cP0000075
|
PotentialSSR
|
HexaSSR
|
33700
|
33711
|
CTCAAG
|
2
| psbC
|
|
cH0000032
|
PotentialSSR
|
PentaSSR
|
33993
|
34002
|
GGTGG
|
2
| psbC
|
|
cP0000076
|
PotentialSSR
|
HexaSSR
|
34118
|
34129
|
TGCAGC
|
2
| psbC
|
|
cP0000077
|
PotentialSSR
|
PentaSSR
|
34316
|
34325
|
TAATT
|
2
| |
|
cH0000033
|
PotentialSSR
|
PentaSSR
|
34411
|
34420
|
AATAA
|
2
| |
|
cP0000078
|
PotentialSSR
|
HexaSSR
|
34560
|
34571
|
TTATTC
|
2
| |
|
cP0000079
|
PotentialSSR
|
PentaSSR
|
34664
|
34673
|
TATAT
|
2
| |
|
cP0000080
|
PotentialSSR
|
PentaSSR
|
35043
|
35052
|
TGGAT
|
2
| psbZ
|
|
cP0000081
|
PotentialSSR
|
PentaSSR
|
35210
|
35219
|
AAACA
|
2
| |
|
cP0000082
|
PotentialSSR
|
PentaSSR
|
35272
|
35281
|
TGAAT
|
2
| |
|
cP0000083
|
PotentialSSR
|
PentaSSR
|
35369
|
35378
|
TATAT
|
2
| |
|
cP0000084
|
PotentialSSR
|
PentaSSR
|
35554
|
35563
|
TAGTG
|
2
| |
|
cP0000085
|
PotentialSSR
|
PentaSSR
|
35579
|
35588
|
TTCTT
|
2
| |
|
cP0000086
|
PotentialSSR
|
PentaSSR
|
36138
|
36147
|
TACTT
|
2
| rps14
|
|
cP0000087
|
PotentialSSR
|
PentaSSR
|
36560
|
36569
|
CCACG
|
2
| psaB
|
|
cP0000088
|
PotentialSSR
|
PentaSSR
|
36709
|
36718
|
CCATC
|
2
| psaB
|
|
cP0000089
|
PotentialSSR
|
PentaSSR
|
37988
|
37997
|
TGTCC
|
2
| psaB
|
|
cH0000034
|
PotentialSSR
|
PentaSSR
|
38494
|
38503
|
ACCAA
|
2
| psaB
|
|
cP0000090
|
PotentialSSR
|
HexaSSR
|
38920
|
38931
|
TAATAG
|
2
| psaA
|
|
cH0000035
|
PotentialSSR
|
PentaSSR
|
39267
|
39276
|
AATGG
|
2
| psaA
|
|
cP0000091
|
PotentialSSR
|
HexaSSR
|
39412
|
39423
|
GTTGTA
|
2
| psaA
|
|
cP0000092
|
PotentialSSR
|
PentaSSR
|
39669
|
39678
|
ATGTG
|
2
| psaA
|
|
cP0000093
|
PotentialSSR
|
PentaSSR
|
40960
|
40969
|
TATTT
|
2
| |
|
cH0000036
|
PotentialSSR
|
PentaSSR
|
41561
|
41570
|
TTTTA
|
2
| |
|
cH0000037
|
PotentialSSR
|
HexaSSR
|
42163
|
42174
|
CTTAGT
|
2
| |
|
cP0000094
|
PotentialSSR
|
HexaSSR
|
42219
|
42230
|
TTATAT
|
2
| |
|
cH0000038
|
PotentialSSR
|
PentaSSR
|
42249
|
42258
|
TTCTT
|
2
| |
|
cP0000095
|
PotentialSSR
|
HexaSSR
|
42332
|
42343
|
CATAGA
|
2
| |
|
cP0000096
|
PotentialSSR
|
PentaSSR
|
43252
|
43261
|
ATTAC
|
2
| |
|
cP0000097
|
PotentialSSR
|
PentaSSR
|
43535
|
43544
|
AAATC
|
2
| |
|
cP0000098
|
PotentialSSR
|
PentaSSR
|
43911
|
43920
|
CAAAT
|
2
| |
|
cP0000099
|
PotentialSSR
|
PentaSSR
|
43978
|
43987
|
GATCA
|
2
| |
|
cP0000100
|
PotentialSSR
|
PentaSSR
|
44511
|
44520
|
GGGAT
|
2
| |
|
cP0000101
|
PotentialSSR
|
PentaSSR
|
44628
|
44637
|
TATTT
|
2
| |
|
cP0000102
|
PotentialSSR
|
PentaSSR
|
45440
|
45449
|
TATAT
|
2
| |
|
cP0000103
|
PotentialSSR
|
PentaSSR
|
45782
|
45791
|
TTTAG
|
2
| |
|
cP0000104
|
PotentialSSR
|
PentaSSR
|
46084
|
46093
|
TAATA
|
2
| |
|
cP0000105
|
PotentialSSR
|
PentaSSR
|
46375
|
46384
|
CAAAT
|
2
| |
|
cP0000106
|
PotentialSSR
|
PentaSSR
|
47143
|
47152
|
TTTTC
|
2
| |
|
cP0000107
|
PotentialSSR
|
PentaSSR
|
47269
|
47278
|
TTATT
|
2
| |
|
cH0000039
|
PotentialSSR
|
PentaSSR
|
47407
|
47416
|
AAATG
|
2
| |
|
cP0000108
|
PotentialSSR
|
HexaSSR
|
47652
|
47663
|
TAAAAT
|
2
| |
|
cH0000041
|
PotentialSSR
|
HexaSSR
|
48538
|
48549
|
CATATA
|
2
| ndhJ
|
|
cH0000042
|
PotentialSSR
|
HexaSSR
|
48698
|
48709
|
TTTGTA
|
2
| |
|
cP0000109
|
PotentialSSR
|
HexaSSR
|
48875
|
48886
|
ATTTAT
|
2
| ndhK
|
|
cP0000110
|
PotentialSSR
|
PentaSSR
|
49572
|
49581
|
TAAAC
|
2
| ndhC
|
|
cH0000044
|
PotentialSSR
|
PentaSSR
|
50095
|
50104
|
TTCCA
|
2
| |
|
cP0000111
|
PotentialSSR
|
HexaSSR
|
50143
|
50154
|
TATACA
|
2
| |
|
cP0000112
|
PotentialSSR
|
PentaSSR
|
50374
|
50383
|
AAAGA
|
2
| |
|
cH0000045
|
PotentialSSR
|
PentaSSR
|
50405
|
50414
|
AGGTA
|
2
| |
|
cH0000046
|
PotentialSSR
|
HexaSSR
|
50472
|
50483
|
TTCAAA
|
2
| |
|
cP0000113
|
PotentialSSR
|
HexaSSR
|
50525
|
50536
|
AGTTAA
|
2
| |
|
cP0000114
|
PotentialSSR
|
PentaSSR
|
50649
|
50658
|
AAATA
|
2
| |
|
cH0000047
|
PotentialSSR
|
PentaSSR
|
51023
|
51032
|
AAATG
|
2
| |
|
cP0000115
|
PotentialSSR
|
HexaSSR
|
51227
|
51238
|
GAACTA
|
2
| |
|
cH0000048
|
PotentialSSR
|
PentaSSR
|
51259
|
51268
|
TTGTT
|
2
| |
|
cH0000049
|
PotentialSSR
|
HexaSSR
|
51738
|
51749
|
TTATTT
|
2
| |
|
cH0000050
|
PotentialSSR
|
HexaSSR
|
51766
|
51777
|
AAAATA
|
2
| |
|
cH0000051
|
PotentialSSR
|
HexaSSR
|
53800
|
53811
|
GAAAAT
|
2
| |
|
cH0000052
|
PotentialSSR
|
HexaSSR
|
53938
|
53949
|
TTCTAT
|
2
| |
|
cP0000116
|
PotentialSSR
|
HexaSSR
|
54216
|
54227
|
ATATAC
|
2
| |
|
cH0000053
|
PotentialSSR
|
PentaSSR
|
54321
|
54330
|
GGTTG
|
2
| |
|
cP0000117
|
PotentialSSR
|
HexaSSR
|
56300
|
56311
|
CCTTTC
|
2
| |
|
cP0000118
|
PotentialSSR
|
PentaSSR
|
56316
|
56325
|
TAGAA
|
2
| |
|
cP0000119
|
PotentialSSR
|
PentaSSR
|
56687
|
56696
|
TTTTC
|
2
| |
|
cP0000120
|
PotentialSSR
|
PentaSSR
|
57949
|
57958
|
GGGTG
|
2
| accD
|
|
cH0000054
|
PotentialSSR
|
PentaSSR
|
58414
|
58423
|
TATAT
|
2
| |
|
cP0000121
|
PotentialSSR
|
HexaSSR
|
58503
|
58514
|
AGAATT
|
2
| |
|
cH0000056
|
PotentialSSR
|
HexaSSR
|
58570
|
58581
|
TACAAT
|
2
| |
|
cP0000122
|
PotentialSSR
|
HexaSSR
|
58905
|
58916
|
TTAGAT
|
2
| |
|
cP0000123
|
PotentialSSR
|
PentaSSR
|
59030
|
59039
|
TTCAA
|
2
| |
|
cP0000124
|
PotentialSSR
|
PentaSSR
|
59340
|
59349
|
TCGAT
|
2
| ycf4
|
|
cP0000125
|
PotentialSSR
|
PentaSSR
|
59445
|
59454
|
TAGAA
|
2
| ycf4
|
|
cP0000126
|
PotentialSSR
|
PentaSSR
|
59656
|
59665
|
GAAAA
|
2
| |
|
cP0000127
|
PotentialSSR
|
PentaSSR
|
59779
|
59788
|
AAATA
|
2
| |
|
cP0000128
|
PotentialSSR
|
PentaSSR
|
59877
|
59886
|
TATAT
|
2
| |
|
cH0000057
|
PotentialSSR
|
PentaSSR
|
59998
|
60007
|
AATTA
|
2
| |
|
cP0000129
|
PotentialSSR
|
HexaSSR
|
60982
|
60993
|
TTTGAA
|
2
| |
|
cP0000130
|
PotentialSSR
|
PentaSSR
|
61025
|
61034
|
ACAAA
|
2
| |
|
cP0000131
|
PotentialSSR
|
PentaSSR
|
61588
|
61597
|
AAAGA
|
2
| petA
|
|
cH0000058
|
PotentialSSR
|
PentaSSR
|
61859
|
61868
|
GAAAA
|
2
| petA
|
|
cH0000059
|
PotentialSSR
|
HexaSSR
|
62195
|
62206
|
TAACAA
|
2
| |
|
cH0000060
|
PotentialSSR
|
HexaSSR
|
62353
|
62364
|
TTGGAT
|
2
| |
|
cP0000132
|
PotentialSSR
|
HexaSSR
|
62578
|
62589
|
ATTTTT
|
2
| |
|
cP0000133
|
PotentialSSR
|
PentaSSR
|
62771
|
62780
|
TTAAC
|
2
| |
|
cP0000134
|
PotentialSSR
|
PentaSSR
|
63243
|
63252
|
GATAA
|
2
| |
|
cP0000135
|
PotentialSSR
|
PentaSSR
|
63260
|
63269
|
TCTAT
|
2
| |
|
cH0000061
|
PotentialSSR
|
PentaSSR
|
63367
|
63376
|
ATTCA
|
2
| psbL
|
|
cP0000136
|
PotentialSSR
|
HexaSSR
|
63421
|
63432
|
ATTCGG
|
2
| |
|
cH0000062
|
PotentialSSR
|
PentaSSR
|
63453
|
63462
|
ATTGC
|
2
| psbF
|
|
cH0000063
|
PotentialSSR
|
HexaSSR
|
64105
|
64116
|
CAAATA
|
2
| |
|
cP0000137
|
PotentialSSR
|
HexaSSR
|
64472
|
64483
|
TTTATA
|
2
| |
|
cP0000138
|
PotentialSSR
|
PentaSSR
|
64613
|
64622
|
TATTT
|
2
| |
|
cP0000139
|
PotentialSSR
|
PentaSSR
|
64924
|
64933
|
TATTT
|
2
| |
|
cP0000140
|
PotentialSSR
|
PentaSSR
|
65382
|
65391
|
GATTA
|
2
| |
|
cP0000141
|
PotentialSSR
|
PentaSSR
|
65502
|
65511
|
GAACT
|
2
| trnW-CCA
|
|
cP0000142
|
PotentialSSR
|
PentaSSR
|
65783
|
65792
|
TTCAA
|
2
| |
|
cP0000143
|
PotentialSSR
|
PentaSSR
|
65822
|
65831
|
CTTGT
|
2
| |
|
cP0000144
|
PotentialSSR
|
PentaSSR
|
65981
|
65990
|
TATCT
|
2
| |
|
cP0000145
|
PotentialSSR
|
PentaSSR
|
66016
|
66025
|
TTTGA
|
2
| |
|
cP0000146
|
PotentialSSR
|
PentaSSR
|
66026
|
66035
|
CTTAG
|
2
| |
|
cP0000147
|
PotentialSSR
|
PentaSSR
|
66354
|
66363
|
TTTTA
|
2
| |
|
cP0000148
|
PotentialSSR
|
PentaSSR
|
66368
|
66377
|
AAATA
|
2
| |
|
cP0000149
|
PotentialSSR
|
PentaSSR
|
66393
|
66402
|
AAAAG
|
2
| |
|
cP0000150
|
PotentialSSR
|
PentaSSR
|
67431
|
67440
|
AAATA
|
2
| rps18
|
|
cP0000151
|
PotentialSSR
|
PentaSSR
|
67552
|
67561
|
AATTG
|
2
| |
|
cP0000152
|
PotentialSSR
|
PentaSSR
|
68695
|
68704
|
TCTTA
|
2
| |
|
cH0000064
|
PotentialSSR
|
HexaSSR
|
68842
|
68853
|
TTTTGT
|
2
| |
|
cP0000153
|
PotentialSSR
|
PentaSSR
|
69499
|
69508
|
CTTTT
|
2
| |
|
cH0000065
|
PotentialSSR
|
HexaSSR
|
69540
|
69551
|
CAGATC
|
2
| |
|
cP0000154
|
PotentialSSR
|
PentaSSR
|
69763
|
69772
|
ATAAA
|
2
| |
|
cH0000066
|
PotentialSSR
|
HexaSSR
|
70123
|
70134
|
ACAAAT
|
2
| clpP
|
|
cP0000155
|
PotentialSSR
|
PentaSSR
|
70528
|
70537
|
TATCA
|
2
| |
|
cP0000156
|
PotentialSSR
|
PentaSSR
|
70543
|
70552
|
ATCCA
|
2
| |
|
cP0000157
|
PotentialSSR
|
PentaSSR
|
70701
|
70710
|
TCTTT
|
2
| |
|
cP0000158
|
PotentialSSR
|
PentaSSR
|
70732
|
70741
|
TATTC
|
2
| |
|
cH0000067
|
PotentialSSR
|
HexaSSR
|
70864
|
70875
|
ATTGGG
|
2
| |
|
cH0000068
|
PotentialSSR
|
HexaSSR
|
70968
|
70979
|
CTAAAA
|
2
| |
|
cP0000159
|
PotentialSSR
|
PentaSSR
|
71313
|
71322
|
GTTTA
|
2
| |
|
cP0000160
|
PotentialSSR
|
PentaSSR
|
71446
|
71455
|
ATAGA
|
2
| |
|
cH0000069
|
PotentialSSR
|
HexaSSR
|
71565
|
71576
|
CATAGT
|
2
| |
|
cH0000070
|
PotentialSSR
|
HexaSSR
|
72083
|
72094
|
GTTTTG
|
2
| psbB
|
|
cP0000161
|
PotentialSSR
|
PentaSSR
|
73357
|
73366
|
CTCTA
|
2
| psbT
|
|
cP0000162
|
PotentialSSR
|
PentaSSR
|
73420
|
73429
|
AAAAT
|
2
| psbT
|
|
cP0000163
|
PotentialSSR
|
PentaSSR
|
74397
|
74406
|
CCTAT
|
2
| |
|
cP0000164
|
PotentialSSR
|
PentaSSR
|
74577
|
74586
|
TAATT
|
2
| |
|
cP0000165
|
PotentialSSR
|
PentaSSR
|
74730
|
74739
|
AAAAT
|
2
| |
|
cP0000166
|
PotentialSSR
|
PentaSSR
|
75511
|
75520
|
ATAGA
|
2
| petB
|
|
cP0000167
|
PotentialSSR
|
PentaSSR
|
75654
|
75663
|
TAACA
|
2
| |
|
cP0000168
|
PotentialSSR
|
PentaSSR
|
75852
|
75861
|
TCTAT
|
2
| |
|
cP0000169
|
PotentialSSR
|
PentaSSR
|
76107
|
76116
|
ATATA
|
2
| |
|
cP0000170
|
PotentialSSR
|
PentaSSR
|
76430
|
76439
|
GAATC
|
2
| |
|
cP0000171
|
PotentialSSR
|
PentaSSR
|
77017
|
77026
|
ATTCA
|
2
| |
|
cP0000172
|
PotentialSSR
|
PentaSSR
|
79136
|
79145
|
TTTCA
|
2
| infA
|
|
cH0000071
|
PotentialSSR
|
HexaSSR
|
80236
|
80247
|
GAAAAA
|
2
| |
|
cH0000072
|
PotentialSSR
|
HexaSSR
|
81084
|
81095
|
AAAAAG
|
2
| |
|
cP0000173
|
PotentialSSR
|
PentaSSR
|
81544
|
81553
|
TTTTA
|
2
| |
|
cP0000174
|
PotentialSSR
|
PentaSSR
|
81556
|
81565
|
TTTTA
|
2
| |
|
cP0000175
|
PotentialSSR
|
PentaSSR
|
82050
|
82059
|
ACCCT
|
2
| rps3
|
|
cP0000176
|
PotentialSSR
|
PentaSSR
|
82287
|
82296
|
CAATT
|
2
| rps3
|
|
cP0000177
|
PotentialSSR
|
PentaSSR
|
82652
|
82661
|
TTTAT
|
2
| rpl22
|
|
cP0000178
|
PotentialSSR
|
PentaSSR
|
83245
|
83254
|
TTTTA
|
2
| |
|
cH0000073
|
PotentialSSR
|
HexaSSR
|
83615
|
83626
|
ATTTTC
|
2
| |
|
cP0000179
|
PotentialSSR
|
PentaSSR
|
84030
|
84039
|
TATGT
|
2
| rpl2
|
|
cP0000180
|
PotentialSSR
|
PentaSSR
|
84378
|
84387
|
TGGAT
|
2
| rpl2
|
|
cP0000181
|
PotentialSSR
|
PentaSSR
|
84479
|
84488
|
TTCTT
|
2
| |
|
cP0000182
|
PotentialSSR
|
PentaSSR
|
84500
|
84509
|
GAATA
|
2
| |
|
cP0000183
|
PotentialSSR
|
PentaSSR
|
84807
|
84816
|
TATGA
|
2
| |
|
cP0000184
|
PotentialSSR
|
PentaSSR
|
85023
|
85032
|
TGAAA
|
2
| |
|
cP0000185
|
PotentialSSR
|
PentaSSR
|
85162
|
85171
|
CAATT
|
2
| ycf2
|
|
cP0000186
|
PotentialSSR
|
PentaSSR
|
85727
|
85736
|
TATAT
|
2
| ycf2
|
|
cP0000187
|
PotentialSSR
|
PentaSSR
|
85741
|
85750
|
GATCC
|
2
| ycf2
|
|
cH0000074
|
PotentialSSR
|
HexaSSR
|
86985
|
86996
|
GAATTT
|
2
| ycf2
|
|
cP0000188
|
PotentialSSR
|
PentaSSR
|
87967
|
87976
|
CGATC
|
2
| ycf2
|
|
cP0000189
|
PotentialSSR
|
PentaSSR
|
88132
|
88141
|
TTCAA
|
2
| ycf2
|
|
cH0000077
|
PotentialSSR
|
HexaSSR
|
89085
|
89096
|
AAGAAA
|
2
| ycf2
|
|
cP0000190
|
PotentialSSR
|
PentaSSR
|
89110
|
89119
|
GATTG
|
2
| ycf2
|
|
cP0000191
|
PotentialSSR
|
PentaSSR
|
90097
|
90106
|
GAAAA
|
2
| ycf2
|
|
cH0000078
|
PotentialSSR
|
HexaSSR
|
90602
|
90613
|
TAGAAG
|
2
| ycf2
|
|
cP0000192
|
PotentialSSR
|
PentaSSR
|
91436
|
91445
|
ATGAA
|
2
| ycf2
|
|
cP0000193
|
PotentialSSR
|
PentaSSR
|
91842
|
91851
|
AAATA
|
2
| |
|
cP0000194
|
PotentialSSR
|
PentaSSR
|
91909
|
91918
|
TTGTT
|
2
| |
|
cH0000079
|
PotentialSSR
|
HexaSSR
|
92140
|
92151
|
ATTCCA
|
2
| |
|
cP0000195
|
PotentialSSR
|
PentaSSR
|
92596
|
92605
|
ATGGA
|
2
| |
|
cH0000080
|
PotentialSSR
|
HexaSSR
|
92774
|
92785
|
CCCATT
|
2
| |
|
cH0000081
|
PotentialSSR
|
HexaSSR
|
93557
|
93568
|
GCTGAA
|
2
| ndhB
|
|
cH0000082
|
PotentialSSR
|
HexaSSR
|
93616
|
93627
|
AGAGTC
|
2
| ndhB
|
|
cP0000196
|
PotentialSSR
|
PentaSSR
|
93697
|
93706
|
TAAGT
|
2
| |
|
cP0000197
|
PotentialSSR
|
PentaSSR
|
93913
|
93922
|
TGATT
|
2
| |
|
cP0000198
|
PotentialSSR
|
PentaSSR
|
94113
|
94122
|
AAAGA
|
2
| |
|
cH0000083
|
PotentialSSR
|
HexaSSR
|
95180
|
95191
|
TTCTTA
|
2
| |
|
cP0000199
|
PotentialSSR
|
PentaSSR
|
95246
|
95255
|
AGAAA
|
2
| |
|
cH0000084
|
PotentialSSR
|
HexaSSR
|
96092
|
96103
|
TCCATA
|
2
| |
|
cP0000200
|
PotentialSSR
|
PentaSSR
|
96280
|
96289
|
CGAAT
|
2
| |
|
cH0000085
|
PotentialSSR
|
HexaSSR
|
96906
|
96917
|
TTCCTC
|
2
| |
|
cH0000086
|
PotentialSSR
|
HexaSSR
|
96918
|
96929
|
TATCCC
|
2
| |
|
cP0000201
|
PotentialSSR
|
PentaSSR
|
97257
|
97266
|
TATTA
|
2
| |
|
cP0000202
|
PotentialSSR
|
PentaSSR
|
97276
|
97285
|
ATTAG
|
2
| |
|
cP0000203
|
PotentialSSR
|
PentaSSR
|
97441
|
97450
|
ATACA
|
2
| |
|
cP0000204
|
PotentialSSR
|
PentaSSR
|
97458
|
97467
|
GCAAT
|
2
| |
|
cP0000205
|
PotentialSSR
|
PentaSSR
|
97492
|
97501
|
GAATG
|
2
| |
|
cH0000088
|
PotentialSSR
|
HexaSSR
|
97583
|
97594
|
TATTAC
|
2
| |
|
cH0000089
|
PotentialSSR
|
HexaSSR
|
97739
|
97750
|
AATGGA
|
2
| |
|
cP0000206
|
PotentialSSR
|
PentaSSR
|
98095
|
98104
|
CAAGA
|
2
| |
|
cP0000207
|
PotentialSSR
|
PentaSSR
|
98179
|
98188
|
AGGGA
|
2
| trnV-GAC
|
|
cH0000090
|
PotentialSSR
|
HexaSSR
|
98303
|
98314
|
GAATGA
|
2
| |
|
cH0000091
|
PotentialSSR
|
HexaSSR
|
99171
|
99182
|
GACACT
|
2
| rrn16
|
|
cP0000208
|
PotentialSSR
|
PentaSSR
|
100203
|
100212
|
GGGGT
|
2
| |
|
cH0000092
|
PotentialSSR
|
HexaSSR
|
100629
|
100640
|
ATGGAA
|
2
| |
|
cH0000093
|
PotentialSSR
|
HexaSSR
|
101258
|
101269
|
AAGAAT
|
2
| |
|
cP0000209
|
PotentialSSR
|
PentaSSR
|
101489
|
101498
|
ACAAA
|
2
| |
|
cP0000210
|
PotentialSSR
|
PentaSSR
|
101808
|
101817
|
TTCAA
|
2
| |
|
cH0000094
|
PotentialSSR
|
HexaSSR
|
103600
|
103611
|
CGCGAG
|
2
| rrn23
|
|
cH0000095
|
PotentialSSR
|
HexaSSR
|
103623
|
103634
|
GAAGCG
|
2
| rrn23
|
|
cP0000211
|
PotentialSSR
|
PentaSSR
|
105110
|
105119
|
GCGGA
|
2
| rrn23
|
|
cH0000096
|
PotentialSSR
|
HexaSSR
|
105521
|
105532
|
TCTATC
|
2
| |
|
cH0000097
|
PotentialSSR
|
HexaSSR
|
105898
|
105909
|
TTCTTA
|
2
| |
|
cH0000098
|
PotentialSSR
|
HexaSSR
|
106326
|
106337
|
CAAGTA
|
2
| |
|
cP0000212
|
PotentialSSR
|
PentaSSR
|
106342
|
106351
|
TAGCA
|
2
| |
|
cP0000213
|
PotentialSSR
|
PentaSSR
|
106366
|
106375
|
GTCAT
|
2
| |
|
cP0000214
|
PotentialSSR
|
PentaSSR
|
106551
|
106560
|
CAGAA
|
2
| |
|
cH0000099
|
PotentialSSR
|
HexaSSR
|
107841
|
107852
|
TAGAAA
|
2
| ycf1
|
|
cH0000100
|
PotentialSSR
|
HexaSSR
|
107908
|
107919
|
TCCTTC
|
2
| ycf1
|
|
cH0000101
|
PotentialSSR
|
HexaSSR
|
107950
|
107961
|
CAAAAT
|
2
| ycf1
|
|
cP0000215
|
PotentialSSR
|
PentaSSR
|
107997
|
108006
|
ACAAA
|
2
| ycf1
|
|
cP0000216
|
PotentialSSR
|
PentaSSR
|
108222
|
108231
|
GAAAT
|
2
| ycf1
|
|
cP0000217
|
PotentialSSR
|
PentaSSR
|
110784
|
110793
|
TTCTA
|
2
| |
|
cP0000218
|
PotentialSSR
|
PentaSSR
|
111211
|
111220
|
ACTTT
|
2
| |
|
cP0000219
|
PotentialSSR
|
PentaSSR
|
111389
|
111398
|
TAAAT
|
2
| |
|
cP0000220
|
PotentialSSR
|
PentaSSR
|
111610
|
111619
|
ATAAG
|
2
| |
|
cP0000221
|
PotentialSSR
|
PentaSSR
|
111716
|
111725
|
TCAAT
|
2
| |
|
cH0000102
|
PotentialSSR
|
HexaSSR
|
111959
|
111970
|
AAGGAT
|
2
| |
|
cH0000103
|
PotentialSSR
|
HexaSSR
|
112079
|
112090
|
TTTTGT
|
2
| |
|
cP0000222
|
PotentialSSR
|
PentaSSR
|
112470
|
112479
|
AGAAA
|
2
| |
|
cP0000223
|
PotentialSSR
|
PentaSSR
|
112665
|
112674
|
TTTCA
|
2
| |
|
cP0000224
|
PotentialSSR
|
PentaSSR
|
112727
|
112736
|
AAATT
|
2
| |
|
cP0000225
|
PotentialSSR
|
PentaSSR
|
112747
|
112756
|
AACAA
|
2
| |
|
cH0000104
|
PotentialSSR
|
HexaSSR
|
112795
|
112806
|
TATGAA
|
2
| |
|
cH0000105
|
PotentialSSR
|
HexaSSR
|
112850
|
112861
|
GAAATG
|
2
| |
|
cP0000226
|
PotentialSSR
|
PentaSSR
|
113017
|
113026
|
GGCAT
|
2
| trnL-UAG
|
|
cP0000227
|
PotentialSSR
|
PentaSSR
|
113085
|
113094
|
AAATA
|
2
| |
|
cP0000228
|
PotentialSSR
|
PentaSSR
|
113385
|
113394
|
TTTCC
|
2
| ccsA
|
|
cP0000229
|
PotentialSSR
|
PentaSSR
|
114312
|
114321
|
AAAGG
|
2
| |
|
cH0000107
|
PotentialSSR
|
HexaSSR
|
114371
|
114382
|
TGAGAG
|
2
| ndhD
|
|
cH0000108
|
PotentialSSR
|
HexaSSR
|
115394
|
115405
|
TAATTC
|
2
| ndhD
|
|
cP0000230
|
PotentialSSR
|
PentaSSR
|
116256
|
116265
|
AGTAA
|
2
| |
|
cP0000231
|
PotentialSSR
|
PentaSSR
|
117129
|
117138
|
TGATT
|
2
| ndhG
|
|
cP0000232
|
PotentialSSR
|
PentaSSR
|
117422
|
117431
|
AATTG
|
2
| ndhG
|
|
cP0000233
|
PotentialSSR
|
PentaSSR
|
117527
|
117536
|
TAAAA
|
2
| |
|
cP0000234
|
PotentialSSR
|
PentaSSR
|
117636
|
117645
|
AAGTA
|
2
| |
|
cH0000110
|
PotentialSSR
|
HexaSSR
|
117800
|
117811
|
AAGAAA
|
2
| |
|
cP0000235
|
PotentialSSR
|
PentaSSR
|
117979
|
117988
|
TAATT
|
2
| ndhI
|
|
cP0000236
|
PotentialSSR
|
PentaSSR
|
118449
|
118458
|
AATTT
|
2
| |
|
cH0000111
|
PotentialSSR
|
HexaSSR
|
118627
|
118638
|
GAACAA
|
2
| ndhA
|
|
cP0000237
|
PotentialSSR
|
PentaSSR
|
118813
|
118822
|
ATAAA
|
2
| ndhA
|
|
cP0000238
|
PotentialSSR
|
PentaSSR
|
119256
|
119265
|
CTTTA
|
2
| |
|
cP0000239
|
PotentialSSR
|
PentaSSR
|
119415
|
119424
|
AAAAT
|
2
| |
|
cP0000240
|
PotentialSSR
|
PentaSSR
|
119518
|
119527
|
TAAAA
|
2
| |
|
cH0000113
|
PotentialSSR
|
HexaSSR
|
120036
|
120047
|
TTCTTA
|
2
| |
|
cP0000241
|
PotentialSSR
|
PentaSSR
|
120937
|
120946
|
TATTT
|
2
| ndhH
|
|
cP0000242
|
PotentialSSR
|
PentaSSR
|
122324
|
122333
|
TTTAT
|
2
| |
|
cH0000114
|
PotentialSSR
|
HexaSSR
|
122599
|
122610
|
AATTTT
|
2
| ycf1
|
|
cP0000243
|
PotentialSSR
|
PentaSSR
|
123021
|
123030
|
TTCTT
|
2
| ycf1
|
|
cH0000115
|
PotentialSSR
|
HexaSSR
|
123896
|
123907
|
TATAAG
|
2
| ycf1
|
|
cH0000116
|
PotentialSSR
|
HexaSSR
|
123949
|
123960
|
CTATAT
|
2
| ycf1
|
|
cH0000117
|
PotentialSSR
|
HexaSSR
|
125650
|
125661
|
TCTATT
|
2
| ycf1
|
|
cH0000118
|
PotentialSSR
|
HexaSSR
|
125727
|
125738
|
TTCTTT
|
2
| ycf1
|
|
cP0000244
|
PotentialSSR
|
PentaSSR
|
125830
|
125839
|
TCTAT
|
2
| ycf1
|
|
cP0000245
|
PotentialSSR
|
PentaSSR
|
126879
|
126888
|
CATTT
|
2
| ycf1
|
|
cP0000246
|
PotentialSSR
|
PentaSSR
|
127105
|
127114
|
TTTGT
|
2
| ycf1
|
|
cH0000119
|
PotentialSSR
|
HexaSSR
|
127149
|
127160
|
GATTTT
|
2
| ycf1
|
|
cH0000120
|
PotentialSSR
|
HexaSSR
|
127192
|
127203
|
GAAGGA
|
2
| ycf1
|
|
cH0000121
|
PotentialSSR
|
HexaSSR
|
127259
|
127270
|
TTTCTA
|
2
| ycf1
|
|
cP0000247
|
PotentialSSR
|
PentaSSR
|
128551
|
128560
|
TTCTG
|
2
| |
|
cP0000248
|
PotentialSSR
|
PentaSSR
|
128736
|
128745
|
ATGAC
|
2
| |
|
cP0000249
|
PotentialSSR
|
PentaSSR
|
128760
|
128769
|
TGCTA
|
2
| |
|
cH0000122
|
PotentialSSR
|
HexaSSR
|
128774
|
128785
|
TACTTG
|
2
| |
|
cH0000123
|
PotentialSSR
|
HexaSSR
|
129201
|
129212
|
ATAAGA
|
2
| |
|
cH0000124
|
PotentialSSR
|
HexaSSR
|
129579
|
129590
|
GATAGA
|
2
| |
|
cP0000250
|
PotentialSSR
|
PentaSSR
|
129992
|
130001
|
TCCGC
|
2
| rrn23
|
|
cH0000125
|
PotentialSSR
|
HexaSSR
|
131477
|
131488
|
CGCTTC
|
2
| rrn23
|
|
cH0000126
|
PotentialSSR
|
HexaSSR
|
131499
|
131510
|
GCTCGC
|
2
| rrn23
|
|
cP0000251
|
PotentialSSR
|
PentaSSR
|
133293
|
133302
|
ATTGA
|
2
| |
|
cP0000252
|
PotentialSSR
|
PentaSSR
|
133613
|
133622
|
TTTGT
|
2
| |
|
cH0000127
|
PotentialSSR
|
HexaSSR
|
133842
|
133853
|
ATTCTT
|
2
| |
|
cH0000128
|
PotentialSSR
|
HexaSSR
|
134466
|
134477
|
TCCATT
|
2
| |
|
cP0000253
|
PotentialSSR
|
PentaSSR
|
134899
|
134908
|
ACCCC
|
2
| |
|
cH0000129
|
PotentialSSR
|
HexaSSR
|
135927
|
135938
|
TCAGTG
|
2
| rrn16
|
|
cH0000130
|
PotentialSSR
|
HexaSSR
|
136797
|
136808
|
TCATTC
|
2
| |
|
cP0000254
|
PotentialSSR
|
PentaSSR
|
136923
|
136932
|
TCCCT
|
2
| trnV-GAC
|
|
cP0000255
|
PotentialSSR
|
PentaSSR
|
137007
|
137016
|
TCTTG
|
2
| |
|
cH0000131
|
PotentialSSR
|
HexaSSR
|
137359
|
137370
|
TTTCCA
|
2
| |
|
cH0000132
|
PotentialSSR
|
HexaSSR
|
137514
|
137525
|
ATAGTA
|
2
| |
|
cP0000256
|
PotentialSSR
|
PentaSSR
|
137610
|
137619
|
CATTC
|
2
| |
|
cP0000257
|
PotentialSSR
|
PentaSSR
|
137643
|
137652
|
CATTG
|
2
| |
|
cP0000258
|
PotentialSSR
|
PentaSSR
|
137661
|
137670
|
TGTAT
|
2
| |
|
cP0000259
|
PotentialSSR
|
PentaSSR
|
137826
|
137835
|
CTAAT
|
2
| |
|
cP0000260
|
PotentialSSR
|
PentaSSR
|
137845
|
137854
|
TAATA
|
2
| |
|
cH0000134
|
PotentialSSR
|
HexaSSR
|
138182
|
138193
|
GGGATA
|
2
| |
|
cH0000135
|
PotentialSSR
|
HexaSSR
|
138194
|
138205
|
GAGGAA
|
2
| |
|
cP0000261
|
PotentialSSR
|
PentaSSR
|
138821
|
138830
|
GATTC
|
2
| |
|
cH0000136
|
PotentialSSR
|
HexaSSR
|
139008
|
139019
|
TATGGA
|
2
| |
|
cP0000262
|
PotentialSSR
|
PentaSSR
|
139856
|
139865
|
TTTCT
|
2
| |
|
cH0000137
|
PotentialSSR
|
HexaSSR
|
139920
|
139931
|
TAAGAA
|
2
| |
|
cP0000263
|
PotentialSSR
|
PentaSSR
|
140989
|
140998
|
TCTTT
|
2
| |
|
cP0000264
|
PotentialSSR
|
PentaSSR
|
141189
|
141198
|
AATCA
|
2
| |
|
cP0000265
|
PotentialSSR
|
PentaSSR
|
141405
|
141414
|
ACTTA
|
2
| |
|
cH0000138
|
PotentialSSR
|
HexaSSR
|
141484
|
141495
|
GACTCT
|
2
| ndhB
|
|
cH0000139
|
PotentialSSR
|
HexaSSR
|
141541
|
141552
|
GCTTCA
|
2
| ndhB
|
|
cH0000140
|
PotentialSSR
|
HexaSSR
|
142326
|
142337
|
AATGGG
|
2
| |
|
cP0000266
|
PotentialSSR
|
PentaSSR
|
142505
|
142514
|
TTCCA
|
2
| |
|
cH0000141
|
PotentialSSR
|
HexaSSR
|
142959
|
142970
|
TTGGAA
|
2
| |
|
cP0000267
|
PotentialSSR
|
PentaSSR
|
143193
|
143202
|
AACAA
|
2
| |
|
cP0000268
|
PotentialSSR
|
PentaSSR
|
143260
|
143269
|
TATTT
|
2
| |
|
cP0000269
|
PotentialSSR
|
PentaSSR
|
143665
|
143674
|
TTTCA
|
2
| ycf2
|
|
cH0000142
|
PotentialSSR
|
HexaSSR
|
144498
|
144509
|
CTTCTA
|
2
| ycf2
|
|
cP0000270
|
PotentialSSR
|
PentaSSR
|
145005
|
145014
|
TTTTC
|
2
| ycf2
|
|
cP0000271
|
PotentialSSR
|
PentaSSR
|
145992
|
146001
|
CAATC
|
2
| ycf2
|
|
cH0000143
|
PotentialSSR
|
HexaSSR
|
146012
|
146023
|
CTTTTT
|
2
| ycf2
|
|
cP0000272
|
PotentialSSR
|
PentaSSR
|
146970
|
146979
|
TTGAA
|
2
| ycf2
|
|
cP0000273
|
PotentialSSR
|
PentaSSR
|
147135
|
147144
|
GATCG
|
2
| ycf2
|
|
cH0000146
|
PotentialSSR
|
HexaSSR
|
148112
|
148123
|
TTCAAA
|
2
| ycf2
|
|
cP0000274
|
PotentialSSR
|
PentaSSR
|
149360
|
149369
|
CGGAT
|
2
| ycf2
|
|
cP0000275
|
PotentialSSR
|
PentaSSR
|
149375
|
149384
|
ATATA
|
2
| ycf2
|
|
cP0000276
|
PotentialSSR
|
PentaSSR
|
149938
|
149947
|
TGAAT
|
2
| ycf2
|
|
cP0000277
|
PotentialSSR
|
PentaSSR
|
150078
|
150087
|
ATTTC
|
2
| |
|
cP0000278
|
PotentialSSR
|
PentaSSR
|
150295
|
150304
|
TCATA
|
2
| |
|
cP0000279
|
PotentialSSR
|
PentaSSR
|
150602
|
150611
|
TATTC
|
2
| |
|
cP0000280
|
PotentialSSR
|
PentaSSR
|
150622
|
150631
|
AAAGA
|
2
| |
|
cP0000281
|
PotentialSSR
|
PentaSSR
|
150723
|
150732
|
AATCC
|
2
| rlp2
|
|
cP0000282
|
PotentialSSR
|
PentaSSR
|
151072
|
151081
|
ACATA
|
2
| rpl2
|
|
cH0000147
|
PotentialSSR
|
HexaSSR
|
151485
|
151496
|
GAAAAT
|
2
| |
Acknowledgments
This research was supported by Rural Development Administration (PJ013855052019).
Author Contributions
Jongsun Park (JP) and Youngjae Chung (YC) designed this research and Yongsung Kim (YK) extract DNA of the sample. JP assembled the chloroplast genome and YK annotate it. YK and JP analyze the chloroplast genomes and all three authors wrote and edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.










