Introduction
Materials and Methods
Plant materials and growth conditions
Photoresponse analysis
Gene expression analysis
Statistical analysis
Results and Discussion
Comparison of growing phenotypes between Bd21 and Bd21-3 lines
Responses to different light conditions of the Bd21 and Bd21-3 lines
Fluence rate responses of the Bd21 and Bd21-3 lines
Analysis of Brachypodium genes whose expression is light-inducible
Introduction
Model plants are important for advances in various areas of plant biology research. So far, Arabidopsis thaliana (hereafter, Arabidopsis) has been widely used as a model plant. However, Arabidopsis is a dicot that is only distantly related to monocots. Thus, a monocot model plant is desirable because monocots include the grasses (for example, Poaceae) that provide the majority of human calories and the sources of biofuels (Keller and Feuillet, 2000; Scholthof et al., 2018). As a monocot model plant, Oryza sativa (rice) has been used with the rapid development of genomic and genetic resources. However, rice is a semi-aquatic tropical grass, so it is not a suitable model plant for small grain cereals such as Triticum aestivum (wheat), Hordeum vulgare (barley) and Avena sativa (oats), and for bioenergy crops such as miscanthus and switchgrass (Brkljacic et al., 2011). These considerations have led to the development of a new model plant for the grasses, and Brachypodium distachyon has been adopted as a monocot model system for the study of temperate cereals and related grasses (Draper et al., 2001; Opanowicz et al., 2008).
B. distachyon (hereafter, Brachypodium) is a small and fast- growing species of grass, having the characteristics of a model plant such as small stature (approximately 20-30 ㎝ at maturity), diploidy (2n = 10), short lifecycle (less than 4 months), small genome (~300 Mb), self-pollination, easy cultivation and transformation (Garvin et al., 2008; Kellogg, 2015; Opanowicz et al., 2008). In addition, Brachypodium is closely related to small grain cereals and bioenergy crops, making it a more appropriate model plant than rice or Arabidopsis. Moreover, high-quality genome sequence has been reported (The International Brachypodium Initiative, 2010), which accelerated a wide range of research to improve grain, forage, and bioenergy crops thus far (Catalan et al., 2014; Scholthof et al., 2018). Accordingly, the use of Brachypodium is rapidly growing, exemplified by the increases in germplasm orders and publications (Brkljacic et al., 2011). Therefore, Brachypodium is widely adopted and used as the monocot model plant.
Light is essential for plant growth and development, providing energy source via photosynthesis and environmental signal to modulate whole life cycle from germination to flowering (Ganesan et al., 2017). Plants monitor external light conditions in terms of quality (i.e., wavelength) and quantity (i.e., fluence rate) using photoreceptors to adjust developmental processes, concordantly with the changing environment. Among plant photoreceptors, red (R) and far-red (FR) photoreceptor phytochromes are the best characterized (Li et al., 2011; Xu et al., 2015). There are two major phytochromes in plants, phytochrome A (phyA) and phytochrome B (phyB). For the control of Arabidopsis seedling de-etiolation, it is well known that phyA exclusively mediate responses to FR light, whereas phyB predominantly mediate responses to R light (Han et al., 2007). In a routine way, in vivo functions of phyA and phyB can be examined by measuring hypocotyl (for dicots) or coleoptile (for monocots) lengths of seedlings grown under FR or R light, respectively. For example, phyA mutant displays elongated hypocotyls under FR light compared with wild-type Arabidopsis plant, while transgenic plant overexpressing phyA exhibits shorter hypocotyls, representing increased responses to light. Similarly, phyB mutant displays elongated hypocotyls under R light, while transgenic plant overexpressing phyB shows shorter hypocotyls than wild-type plant. Thus, plant responses to light can be investigated by measuring seedling growths under different light conditions.
Although responses to light have been well characterized in Arabidopsis, only limited information is available for monocots. In Brachypodium, it has been shown that B. distachyon phytochrome C (BdphyC) plays an essential role in photoperiodic flowering from the study of loss-of-function mutants of BdphyC (Woods et al., 2014). However, there is no report of phyA or phyB, including photoresponses of Brachypodium plants to red and far-red light. Thus, we set out to explore responses to light in B. distachyon using two widely-used inbred lines, Bd21 and Bd21-3. The Bd21 is the community standard line that has been used for genome sequencing (The International Brachypodium Initiative, 2010), and the Bd21-3 has been known as a readily transformable line and used for the generation of T-DNA insertional mutants (Tyler et al., 2014; Vogel and Hill, 2008). In this study, we first compared the growing phenotypes between the Bd21 and Bd21-3 lines, and then investigated seedling growths of the Brachypodium lines under different light conditions including dark, far-red, red, and white light. In addition, we examined light-induced expression of three light- responsive genes in Brachypodium. Through this study, we could provide the conditions for photoresponse analysis of the monocot model plant and the reference genes for the future light signaling studies.
Materials and Methods
Plant materials and growth conditions
B. distachyon inbred lines Bd21 and Bd21-3 were obtained from Dr. David Garvin in the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) and Prof. Chung-Mo Park in Seoul National University, respectively. Brachypodium plants were grown in a culture room with 18 h light/6 h dark photoperiod, fluence rate of 150 μmol・m-2・s-1, and temperature of 21-23°C. For analysis of growth stages (from germination to senescence), Brachypodium seeds were sown and grown in 40-cell trays (400 × 270 × 150 ㎜) and 1 plant per cell with soil containing perlite, vermiculite, and humus (in the ratio of 1:2:2). Plants were watered every 3-4 days by sub-irrigation and growth stages were recorded from the day of seeds sown. In our growth conditions, the Brachypodium seeds were germinated in 7 days. Plant stages, plant heights, and number of tillers per plant were investigated as described previously (Hong et al., 2011).
Photoresponse analysis
For measurements of coleoptile and primary leaf lengths, lemmas of mature Bd21 and Bd21-3 seeds were removed and surface-sterilized in 70% (v/v) ethanol for 5 min and 2% (w/v) sodium hypochlorite for 10 min with gently shaking. Seeds were then rinsed several times with sterile distilled water, and the surface-sterilized seeds were placed on half strength Murashige and Skoog (MS) medium containing 0.8% phytoagar, pH 5.8. After 7 days of cold/dark treatment, the seeds were exposed to white light for 8 h at 21°C to enhance germination. For the experiments with continuous light irradiation, the seeds were grown under continuous darkness (D), or kept under D for 1 day and then exposed to far-red (FR, λmax = 738 ㎚, bandwidth = 45 ㎚; 5 μmol・m-2・s-1) or red (R, λmax = 654 ㎚, bandwidth = 25 ㎚; 5 μmol・m-2・s-1) light using LED plant growth chambers (Vision Korea), or white light (WL, 150 μmol・m-2・s-1) for 2-12 days. For the investigation of fluence rate-response curves, seedlings were grown for 7 days under FR or R light with various fluence rates (0~10 μmol・m-2・s-1). The photometer (International Light, USA; Model IL1700) was used to measure the light intensity. After light treatment, seedlings were taken pictures and Scion Image program (http://www.scioncorp.com, Frederick, MD, USA) was used to measure the lengths of coleoptiles and primary leaves, as described previously (Takano et al., 2001).
Gene expression analysis
To investigate the transcript levels of light-responsive genes upon exposure to FR or R light in a time-dependent manner, Bd21 seedlings were grown in the dark for 7 days and harvested at 0, 4, 12, and 24 h after FR or R light treatment (fluence rate = 10 μmol・m-2・s-1). We selected three genes based on Arabidopsis light-responsive genes: chlorophyll A/B binding protein (CAB), light-harvesting chlorophyll binding protein (Lhcb) and chalcone synthase (CHS), which corresponds to BdCAB (Bradi4g37210), BdLhcb (Bradi1g15290), and BdCHS (Bradi4g17230). Total RNA was prepared using RNeasy Plant Mini Kit (Qiagen, Germany) including DNaseI digestion to remove contaminated genomic DNA. The cDNA synthesis was conducted using RNA to cDNA EcoDryTM Premix (Clontech, Japan), and quantitative RT-PCR was performed with Brilliant III UltraFast SYBR® Green QPCR Master Mix (Agilent Technologies, USA) with Stratagene MX3000P according to manufacturer’s instructions. For the analysis of quantitative gene expression, the provided software (MxPro-Mx3000P v.4.10Build 389, Schema85) was used. All tested genes were triplicate and the following primers were used:
5′-AGCTCGAGGTCATCCACTGC-3′ (forward, for BdCAB)
5′-TGTTGTTCACTGGGTCGGAC-3′ (reverse, for BdCAB)
5′-CACCAGACCACCAGCTTCCT-3′ (forward, for BdLhcb)
5′-GCCTCCACCAACACGGTAAC-3′ (reverse, for BdLhcb)
5′-TGTTCCGAGATCACAGCAGTG-3′ (forward, for BdCHS)
5′-GAGGATGAAGAGCACGCAGG-3′ (reverse, for BdCHS)
In addition, gene expression of BdUBQ10 (Bradi1g32860; Chambers et al., 2012) gene was also analysed for normalization by using the primers, 5′-TCCACACTCCACTTGGTGCT-3′ (forward) and 5′-GAGGGTGGACTCCTTTTGGA-3′ (reverse).
Statistical analysis
IBM SPSS statistics 20 software was used to perform ANOVA to analyse the results of the physiological parameters. A significant difference from the control value was determined at P < 0.05 (marked with *) or P < 0.001 level (marked with **). All data represented the mean ± standard deviation (SD) or standard error (SE) of at least three independent experiments.
Results and Discussion
Comparison of growing phenotypes between Bd21 and Bd21-3 lines
The growth stages of the Brachypodium lines were investigated according to the previous report (Hong et al., 2011). In our culture conditions, both seeds were germinated in 7 days and secondary leaves were formed at 14 or 15 days for Bd21 and Bd21-3, respectively (Fig. 1A). Subsequent leaf development overlapped with the stages of tillering and stem elongation, in which the tillering was observed at 21 and 23 days for Bd21 and Bd21-3, respectively. Thus, the growth stages from germination to tillering were very similar between Bd21 and Bd21-3 lines. However, the reproductive growth stages were quite different. The stem elongation stage was through with the appearance of the flag leaf, and emergence of the head at the tip of flag leaf represented the start of the reproductive growth stages. When we compared the heading and flowering stages, the Bd21-3 plants showed much more delayed heading and flowering (55 and 71 days) than the Bd21 plants (38 and 48 days). These differences affected on other growth stages, including seed development, ripening, and senescence. Accordingly, the Bd21-3 plants were taller and have more tillers than the Bd21 plants (Fig. 1B and 1C). These results suggest that the Bd21 line might be a better model plant than the Bd21-3 line, based on the plant height (28.8 vs. 42.4 ㎝) and growing time to harvest seeds (64 vs. 87 days for ripening). However, the Bd21-3 line has been reported to be readily transformed by Agrobacterium-mediated method and to show a low seed dormancy (Barrero et al., 2012; Vogel and Hill, 2008), the Bd21-3 line might be better for genetic transformation. In addition, our results suggest that the experiments with early growth stages might be performed similarly between the two lines, such as photoresponse analysis using seedlings. Thus, we further explored responses to light using the seedlings of Bd21 and Bd21-3 lines.
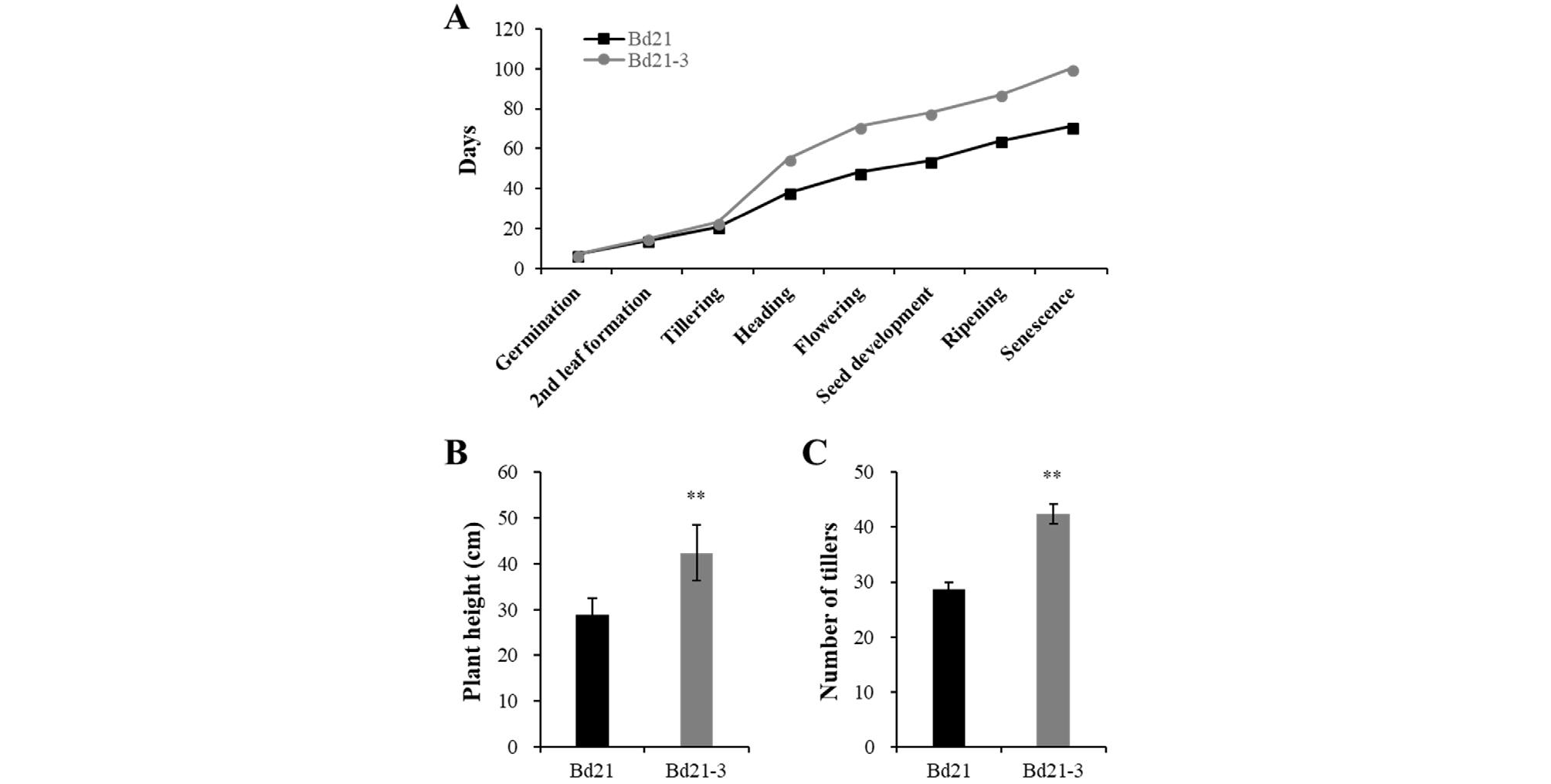
Fig. 1.
Comparison of growing phenotypes between two varieties of Brachypodium distachyon, Bd21 and Bd21-3. (A) Progression of growth stages under long day growth condition (18 h light /6 h dark cycle). (B-C) Plant heights and tiller numbers from fully grown plants. Data represent means ± SE (n = 12). The data showing a statistically significant difference compared with the other data (at P < 0.001) are indicated by **.
Responses to different light conditions of the Bd21 and Bd21-3 lines
To gain basic information about photoresponses of Brachypodium, we investigated seedlings of the Bd21 and Bd21-3 lines grown under continuous dark (D), far-red (FR), red (R), and white light (WL) conditions for 12 days. First, dark-grown seedlings of both Bd21 and Bd21-3 lines exhibited a typical etiolated phenotype such as elongated coleoptiles, absence of chlorophylls (i.e., yellowish leaves), and lack of secondary leaf development (Fig. 2A). In the dark, coleoptiles elongated rapidly and reached at the saturated length around 6 to 8 days (Fig. 2B). In contrast, primary leaves elongated continuously during the growth period (Fig. 2C). The primary leaf lengths could be saturated after 12 days, but the lengths were longer than the square dish that we used for seedling growth, which made it impossible to measure the length correctly. Thus, these results suggest that the coleoptile length is a better parameter to analyze photoresponses of Brachypodium plants than the primary leaf length. Moreover, our results indicate that the coleoptile lengths at approximately 7th day are enough to represent the responses to light in further analysis.
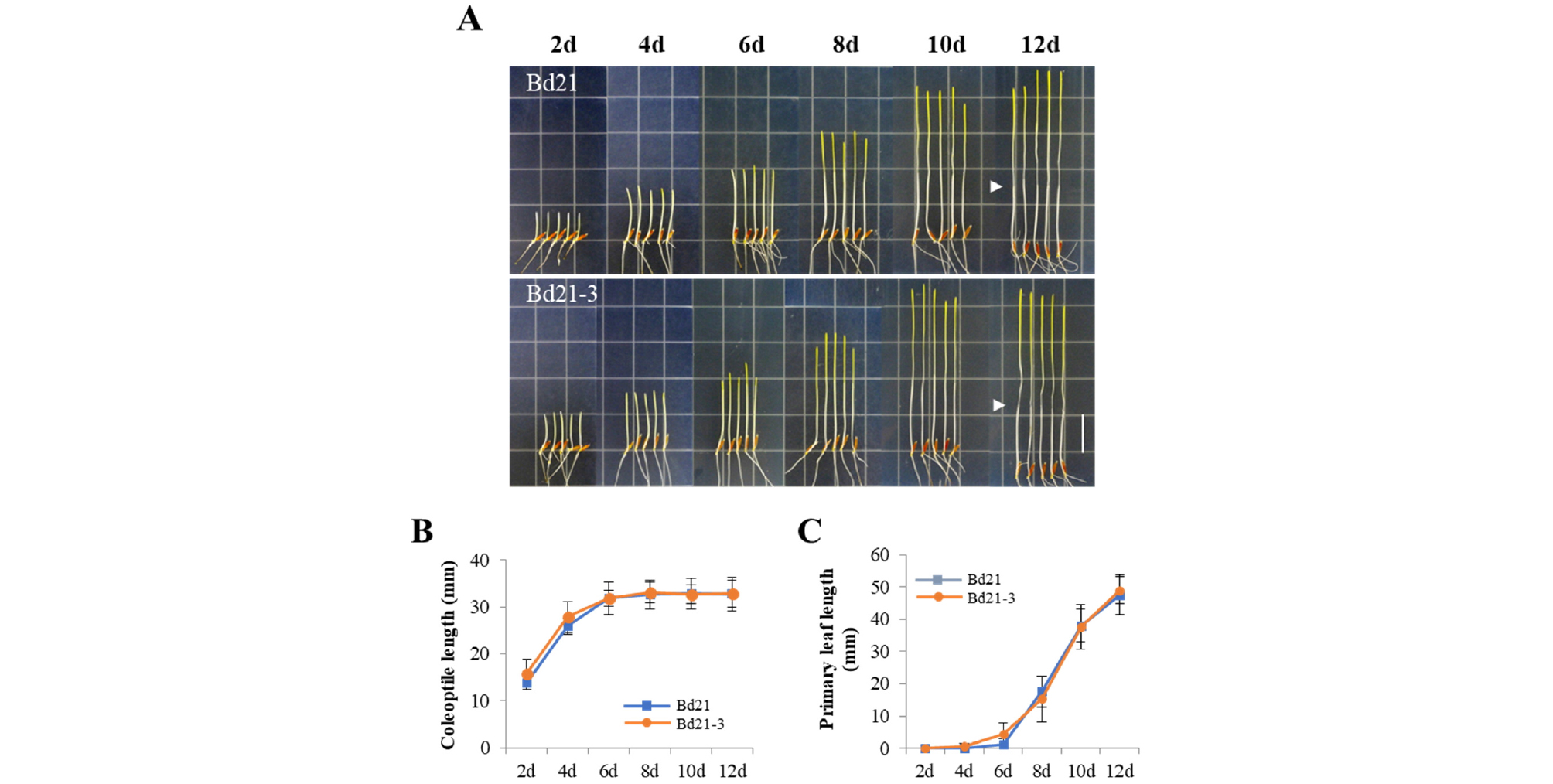
Fig. 2.
Seedling growth of Bd21 and Bd21-3 under dark condition. (A) Representative seedlings grown under dark condition over the course of growth time indicated (2-12 days). Arrowheads indicate the end of coleoptiles at 12th day. Scale bar = 18 ㎜. (B-C) Measurements of coleoptile and primary leaf lengths. Each value indicates the mean ± SD (n ≥ 15).
Next, we investigated the seedlings grown under FR light condition. In this light condition, because photomorphogenic responses can be induced by phyA, we observed inhibition of coleoptile elongation and slightly greened leaves (Fig. 3A). The coleoptile length was saturated to approximately 16 ㎜ after 4 days’ growth (Fig. 3B), which is much shorter than that in the dark (~32 ㎜). Again, primary leaves elongated continuously during the growth period in the FR light condition (Fig. 3C). In our experimental conditions, there was no difference in the results of both Bd21 and Bd21-3 seedlings under FR light.
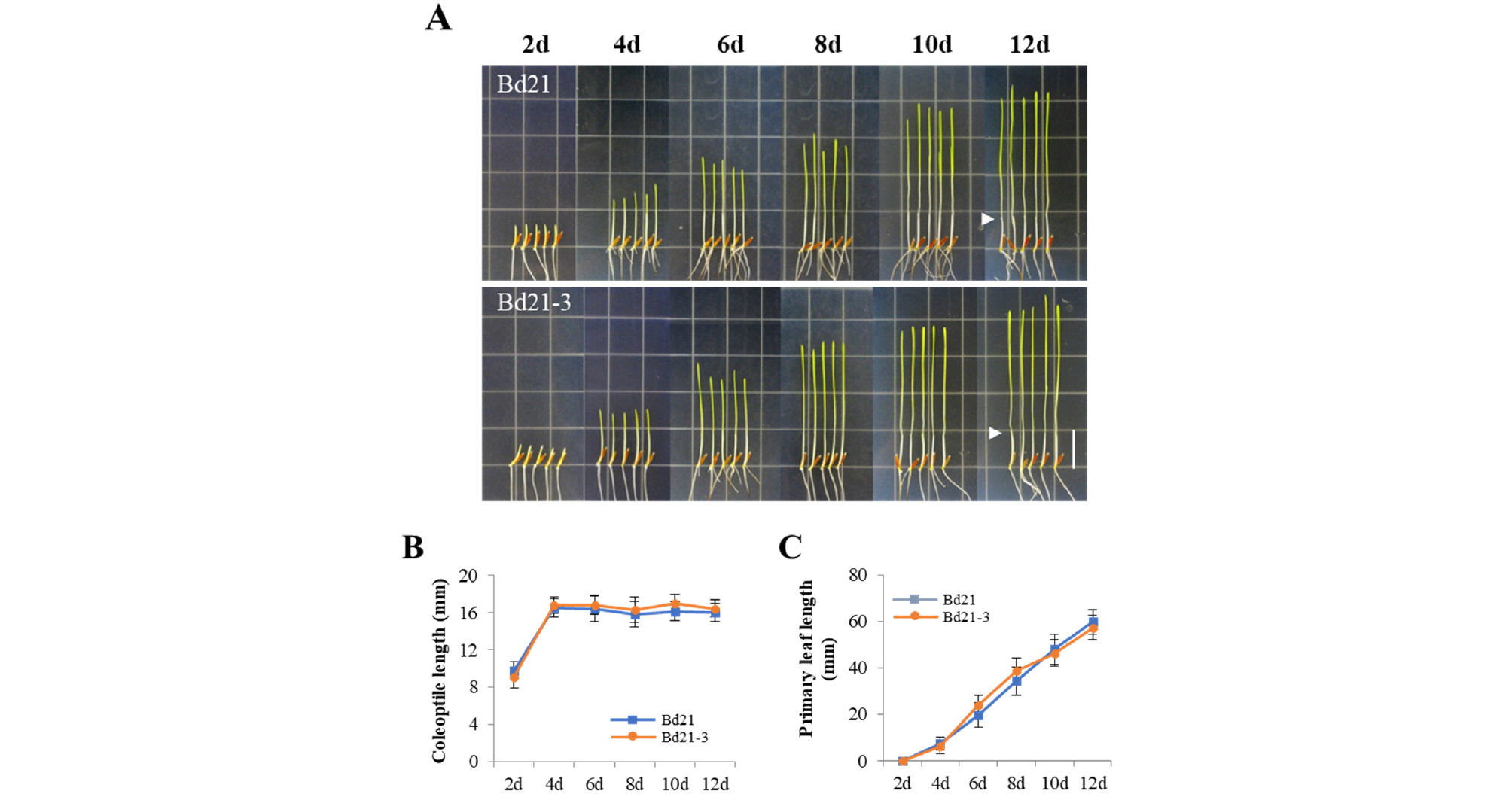
Fig. 3.
Seedling growth of Bd21 and Bd21-3 under continuous far-red light. (A) Representative seedlings grown under far-red light condition (FR, 5 μmol・m-2・s-1) over the course of growth time indicated (2-12 days). Arrowheads indicate the end of coleoptiles at 12th day. Scale bar = 18 ㎜. (B-C) Measurements of coleoptile and primary leaf lengths. Each value indicates the mean ± SD (n ≥ 15).
Under the R light condition, we observed clear photomorphogenic responses, not only inhibition of coleoptile elongation but also accumulation of chlorophylls (i.e., green leaves) and secondary leaf development (Fig. 4A). Similar to the FR light condition, the coleoptile length was saturated after 4 days’ growth (Fig. 4B), which is slightly shorter than that in the FR light (~14/15 ㎜ in R vs. ~16/17 ㎜ in FR for Bd21/Bd21-3). In the case of primary leaves, they emerged earlier than D and FR light conditions, so the growth was saturated after 10 or 12 days’ growth for Bd21-3 and Bd21, respectively (Fig. 4C). Subsequently, we also observed the secondary leaf development, in which the Bd21-3 seedlings formed the second leaves faster than the Bd21 seedlings (for example, ~80% of total Bd21-3 seedlings vs. ~50% of total Bd21 seedlings at 12th day). These results suggest that the R light is enough to induce strong photoresponses including leaf development, compared with the FR light condition.
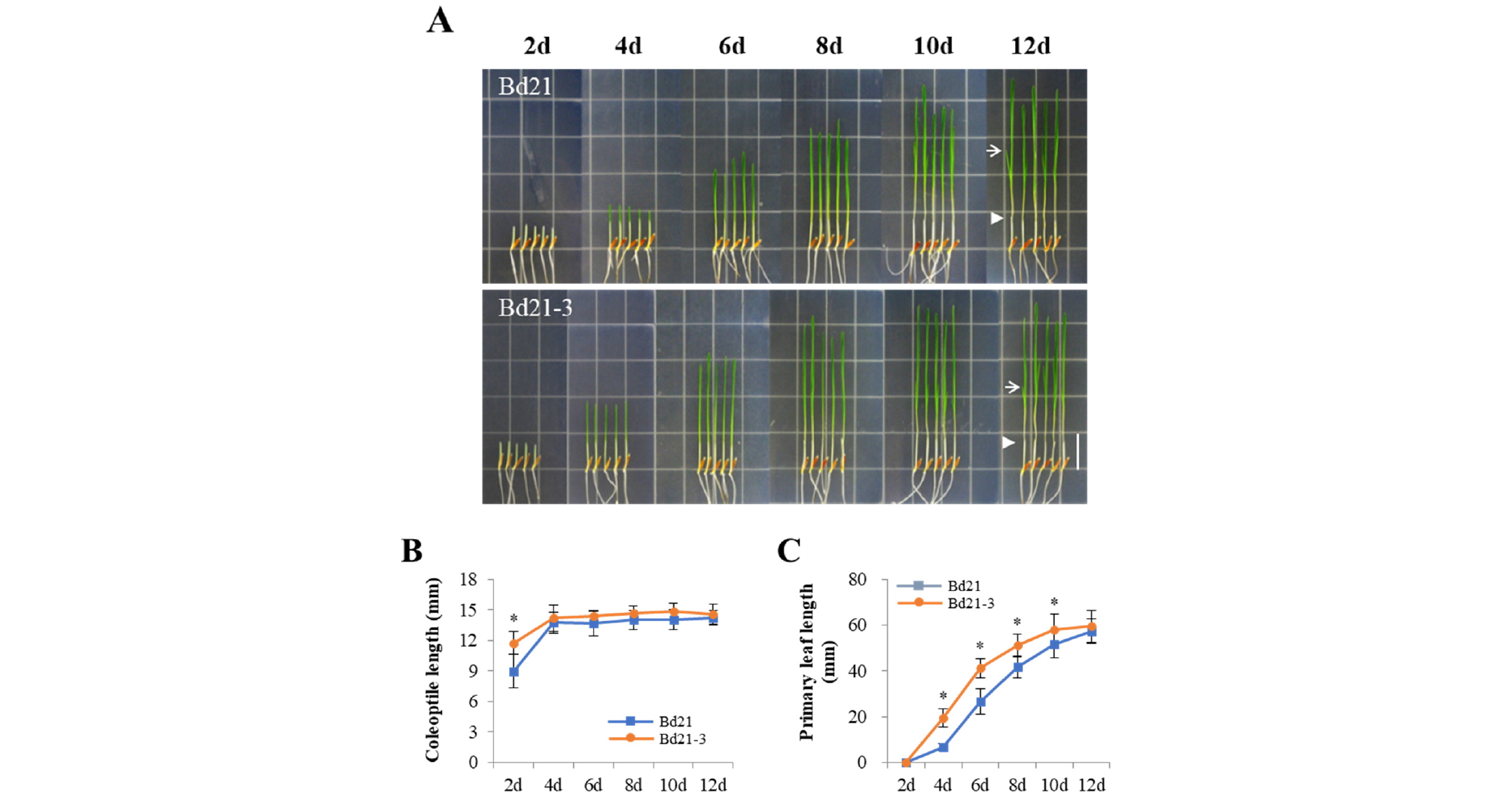
Fig. 4.
Seedling growth of Bd21 and Bd21-3 under continuous red light. (A) Representative seedlings grown under continuous red light condition (R, 5 μmol・m-2・s-1) over the course of growth time indicated (2-12 days). Arrowheads and arrows indicate the end of coleoptiles and the secondary leaves at 12th day, respectively. Scale bar = 18 ㎜. (B-C) Measurements of coleoptile and primary leaf lengths. Each value indicates the mean ± SD (n ≥ 15). The data showing a statistically significant difference (at P < 0.05) are indicated by *.
We also investigated the seedlings grown under WL condition. The growth and development of the Bd21-3 seedlings were rapid compared with those of the Bd21 seedlings (Fig. 5A). In this condition, the Bd21 seedlings produced a shorter coleoptile (~11 ㎜) than the Bd21-3 seedlings (~14.5 ㎜), in which coleoptile lengths of both seedlings were saturated within 4 days’ growth (Fig. 5B). When the primary leaf lengths were compared, the Bd21-3 seedlings were longer than the Bd21 seedlings (Fig. 5C). This is because the primary leaves emerged very early (within 2 days after germination). The secondary leaves of the Bd21 and Bd21-3 seedlings emerged at similar time (around 6 days), but the secondary leaves of the Bd21-3 seedlings were developed much faster than those of the Bd21 seedlings (Fig. 5A). Thus, we could observe the third leaves at the tip of main culm earlier in Bd21-3 (at 10th day) than in Bd21 (at 12th day). In addition, we investigated leaf angles between the main culm and the secondary leaves. The results showed that the Bd21 seedlings exhibited the angles slightly larger than the Bd21-3 seedlings until 8th day (see data of 7 and 8 days in Fig. 5D), but the angles were much larger in the Bd21-3 seedlings after 8 days than the Bd21 seedlings (see data of 10 and 12 days in Fig. 5D). These results also suggest the rapid growth behavior of Bd21-3 compared with Bd21. Collectively, while the Bd21 and Bd21-3 seedlings displayed similar growth patterns in D and FR light conditions, Bd21-3 seedlings showed a rapid growth and development in R and WL conditions. In addition, our results also suggest the Bd21 line as a better model monocot plant for photoresponse analysis, because the seedlings exhibited shorter coleoptiles in all the light conditions tested. When we compared coleoptile lengths in the dark (~32 ㎜) with those under different light conditions, Bd21 seedlings showed the shortest coleoptiles under WL condition (~10 ㎜), followed by R (~14 ㎜) and FR (~16 ㎜) light conditions.
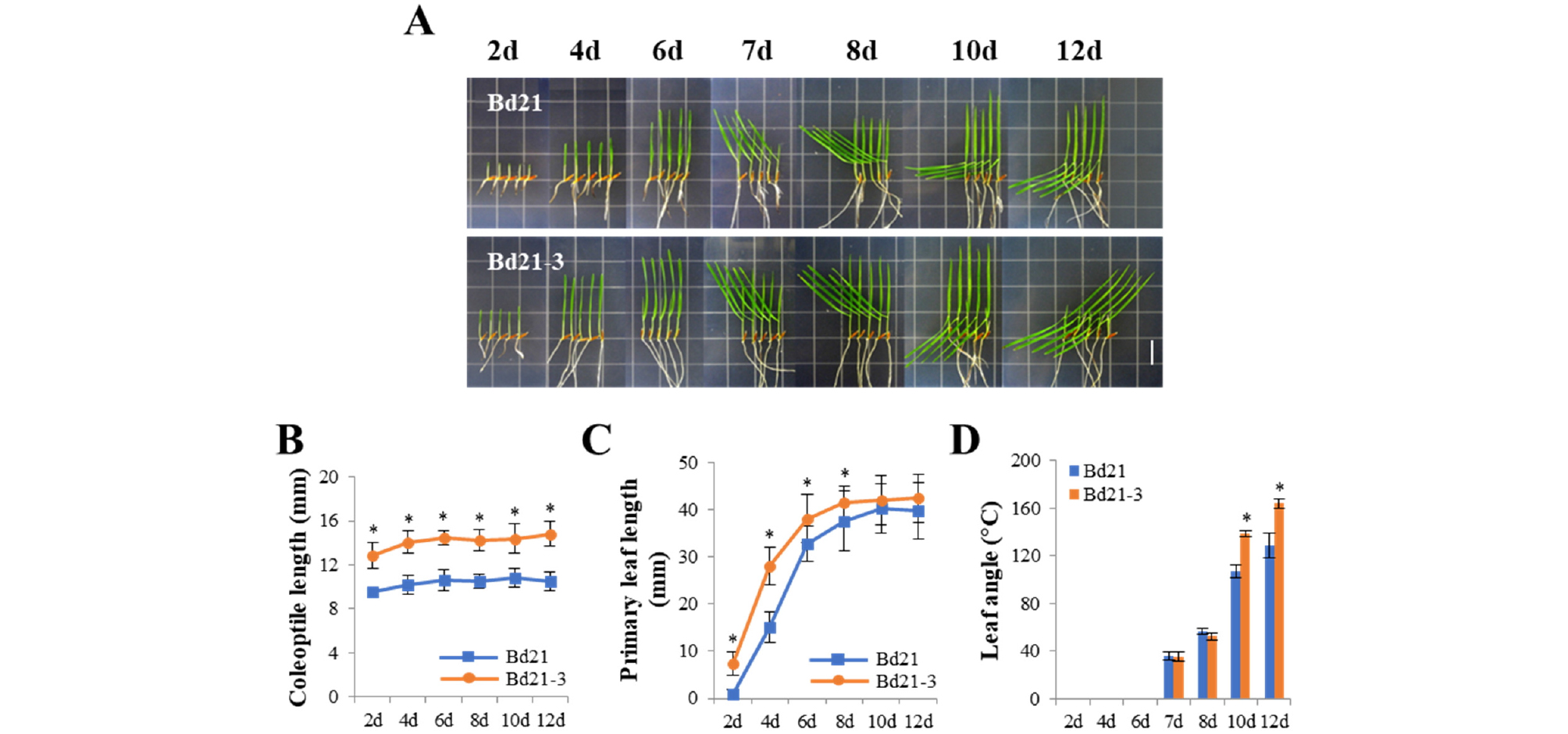
Fig. 5.
Seedling growth of Bd21 and Bd21-3 under continuous white light. (A) Representative seedlings grown under continuous white light condition (WL, 150 μmol・m-2・s-1) over the course of growth time indicated (2-12 days). Scale bar = 18 ㎜. (B-D) Measurements of coleoptile lengths, primary leaf lengths, and angles between monocotyledon and primary leaf. Each value indicates the mean ± SD (n ≥ 15). The data showing a statistically significant difference (at P < 0.05) are indicated by *.
Fluence rate responses of the Bd21 and Bd21-3 lines
For the photoresponse analysis of Arabidopsis, hypocotyl lengths are routinely measured in a fluence rate-dependent manner (Fankhauser and Casal, 2004). Thus, we observed coleoptile lengths under different fluence rates of FR and R light. Treatment of the Bd21 and Bd21-3 seedlings with both light conditions induced inhibition of coleoptile elongation at all fluence rates tested (Fig. 6). The degree of inhibition correlated with the increases in fluence rates applied (i.e., shorter coleoptile lengths with increasing fluence rates). The responses to FR and R light were observed from 0.01 μmol・m-2・s-1 and became saturated around 1 μmol・m-2・s-1. At higher fluence rates (1 and 10 μmol・m-2・s-1), coleoptile elongation was inhibited nearly 50% in both FR and R light conditions (Fig. 6C and 6D). In our experimental conditions, there was no difference in data of the Bd21 and Bd21-3 lines. Overall, our results indicate that, similar to the results of Arabidopsis, fluence-rate response curves could be obtained in the monocot model plant by measuring coleoptile lengths under difference light intensities.
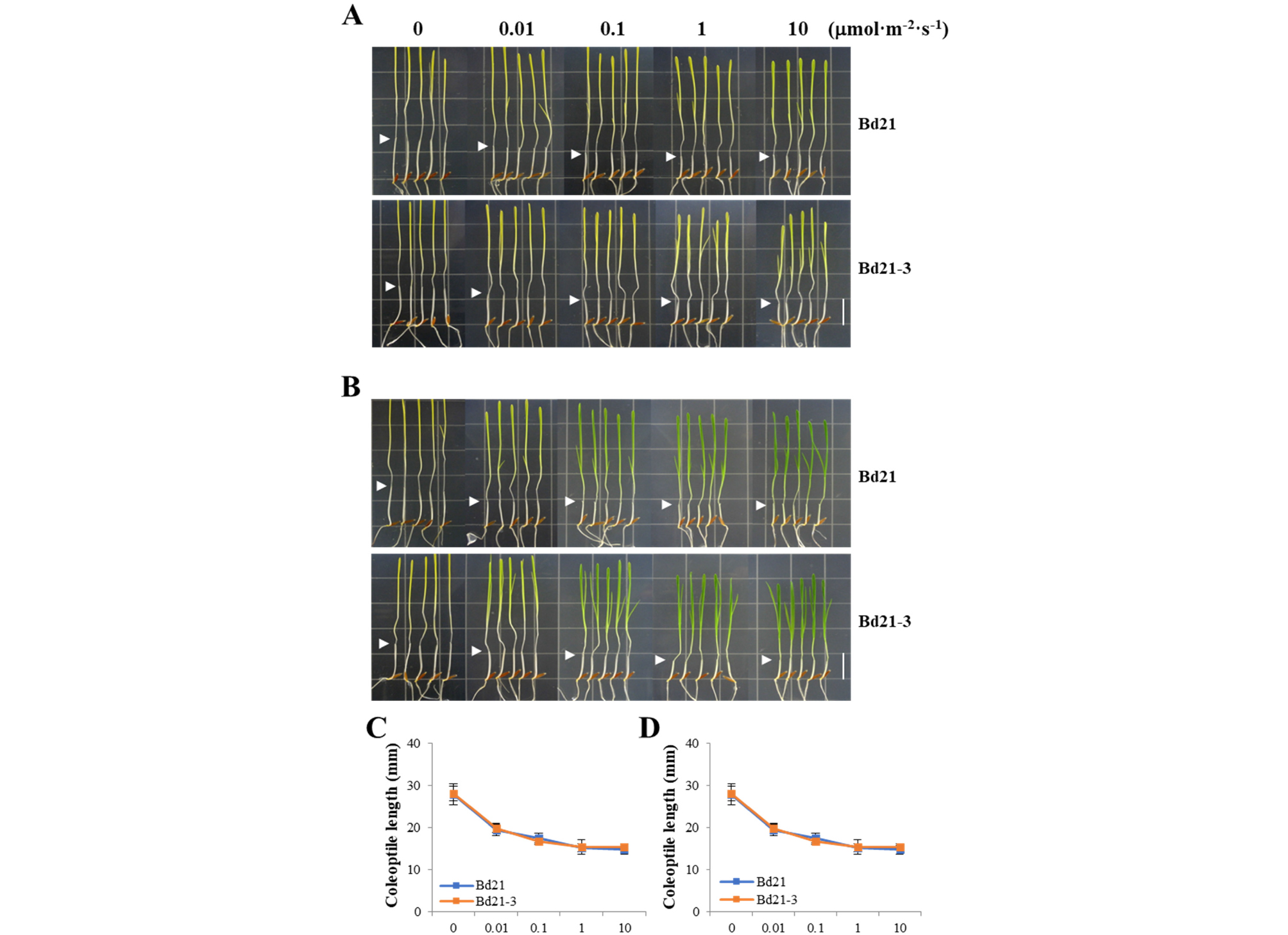
Fig. 6.
Fluence rate-response curves of Bd21 and Bd21-3 under far-red and red light. (A-B) Representative seedlings grown under different far-red (A) and red (B) fluence rates. Arrowheads indicate the end of coleoptiles. Scale bar = 18 ㎜. (C-D) Measurements of coleoptile lengths in (A) and (B), respectively. Each value indicates the mean ± SD (n ≥ 24).
Analysis of Brachypodium genes whose expression is light-inducible
One of quick photoresponses in plants might be expression of light-responsive genes. In Arabidopsis, several light-responsive genes have been used to represent photoresponses to a specific light condition (Shin et al., 2016). Thus, we tried to obtain a few reference genes to represent photoresponses in Brachypodium. For this, we first selected well-known light-responsive genes in Arabidopsis and then found the corresponding Brachypodium genes. In this study, we included genes for chlorophyll A/B binding protein (CAB), light-harvesting chlorophyll binding protein (Lhcb), and chalcone synthase (CHS). Thus, we investigated the expression of these genes in Brachypodium, BdCAB (Bradi4g37210), BdLhcb (Bradi1g15290), and BdCHS (Bradi4g17230), in response to FR and R treatment. The results showed that these genes were all induced by FR and R light in a time-dependent manner (Fig. 7). The transcript levels of BdCAB and BdLhcb were gradually increased in R light treatment, whereas those were increased drastically at 24 h after FR treatment. The magnitudes in increases of BdCAB and BdLhcb expression were similar in both FR and R treatments. These results suggest that both BdCAB and BdLhcb can be used as reference genes for responses to either FR or R light. In the case of BdCHS, the expression was increased gradually in FR light treatment, while it was highly expressed at 12 h after R light treatment (~400 fold in 12 h). The magnitude in the increase of BdCHS expression was much higher in R light treatment than in FR treatment. Thus, the expression of BdCHS can be used as an indicator for the response to R light.
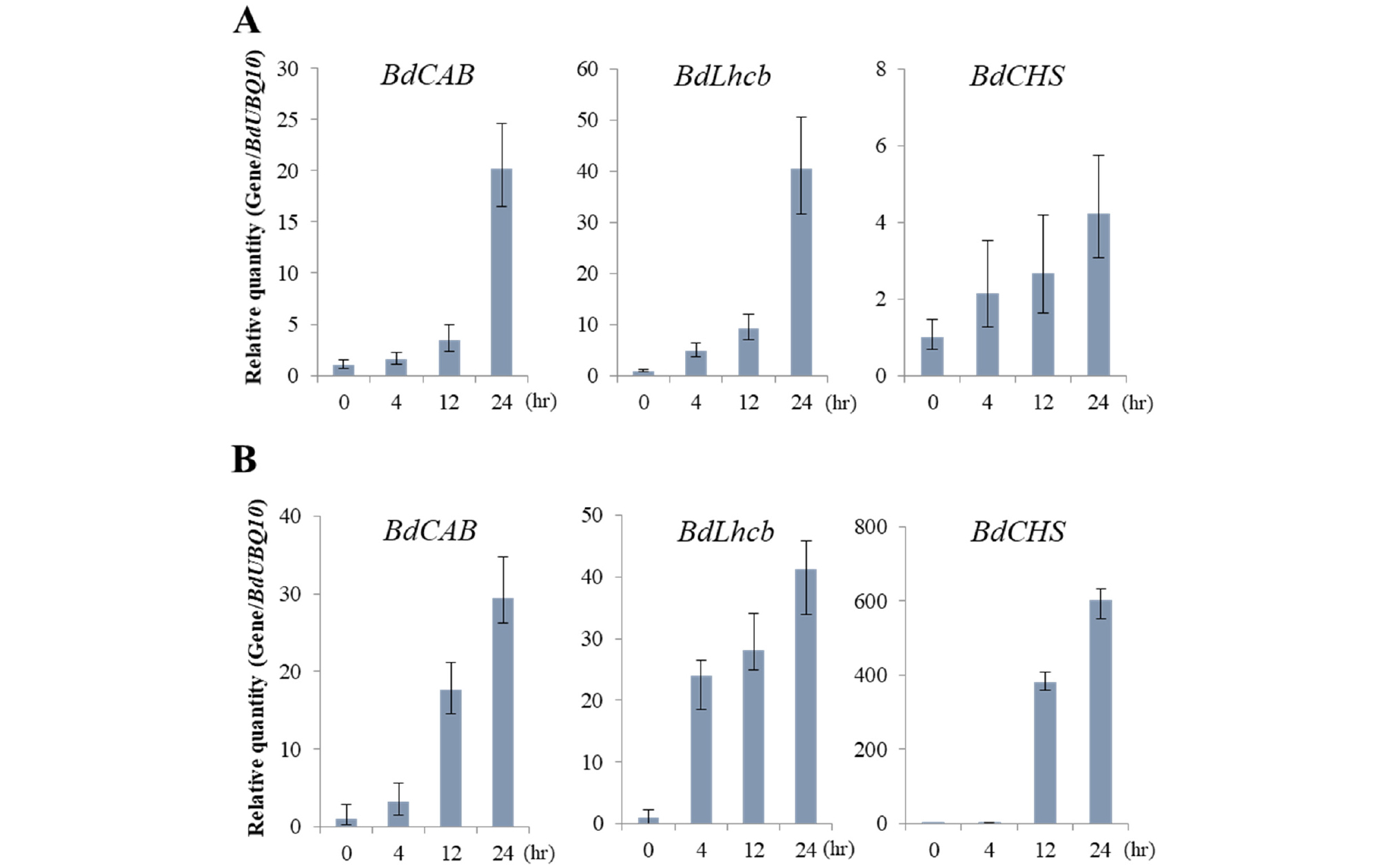
Fig. 7.
Light-induced gene expression analysis in Bd21. Gene expression in response to far-red (A) and red (B) light was analyzed by real-time PCR (n = 3). Bd21 seedlings were grown in the dark for 7 days, and then exposed to FR (10 μmol・m-2・s-1) or R (10 μmol・m-2・s-1) for the time indicated. The gene expression of B. distachyon chlorophyll A/B binding protein (BdCAB), light-harvesting chlorophyll A/B protein (BdLhcb) and chalcone synthase (BdCHS) was normalized to that of BdUBQ10.
In conclusion, appropriate responses to light are crucial for plant growth, development, and survival. Thus, understanding photoresponses of plants might be a basic step to perform plant biology research. Recently, Brachypodium is widely studied as an attractive monocot model plant, because it is a relative of temperate grasses and bioenergy crops. In this regard, we explored responses of Brachypodium plants to different light conditions. In addition, we included two widely used Brachypodium inbred lines, Bd21 and Bd21-3, to choose a better line for future photoresponse analysis. As a result, we could suggest the Bd21 line as a better model to investigate photoresponses in Brachypodium, because it exhibited shorter height, less time taken to harvest seeds, shorter coleoptiles and faster saturated inhibition of coleoptile elongation in response to light treatment. Our data also showed that the coleoptile length can be used as a clear indication for photoresponses, compared with the primary leaf length. In addition, the coleoptile lengths at 7th day after treatment of light conditions provide stable quantitative data to represent photoresponses of Brachypodium plants. Moreover, the present study also provides three reference genes to represent photoresponses in Brachypodium. Collectively, we established the experimental conditions and methods for the photoresponse analysis in Brachypodium, which can be used for the light signaling studies such as phytochrome-mediated photomorphogenesis.




